Main Menu (Mobile)- Block
- Overview
-
Support Teams
- Overview
- Anatomy and Histology
- Cryo-Electron Microscopy
- Electron Microscopy
- Flow Cytometry
- Gene Targeting and Transgenics
- High Performance Computing
- Immortalized Cell Line Culture
- Integrative Imaging
- Invertebrate Shared Resource
- Janelia Experimental Technology
- Mass Spectrometry
- Media Prep
- Molecular Genomics
- Primary & iPS Cell Culture
- Project Pipeline Support
- Project Technical Resources
- Quantitative Genomics
- Scientific Computing
- Viral Tools
- Vivarium
- Open Science
- You + Janelia
- About Us
Main Menu - Block
- Overview
- Anatomy and Histology
- Cryo-Electron Microscopy
- Electron Microscopy
- Flow Cytometry
- Gene Targeting and Transgenics
- High Performance Computing
- Immortalized Cell Line Culture
- Integrative Imaging
- Invertebrate Shared Resource
- Janelia Experimental Technology
- Mass Spectrometry
- Media Prep
- Molecular Genomics
- Primary & iPS Cell Culture
- Project Pipeline Support
- Project Technical Resources
- Quantitative Genomics
- Scientific Computing
- Viral Tools
- Vivarium
How does an animal learn, store, and recall specific items and events? We study spatial learning in rats and mice with a focus on the hippocampus, which has been shown to be a critical brain area underlying spatial behavior and memory.
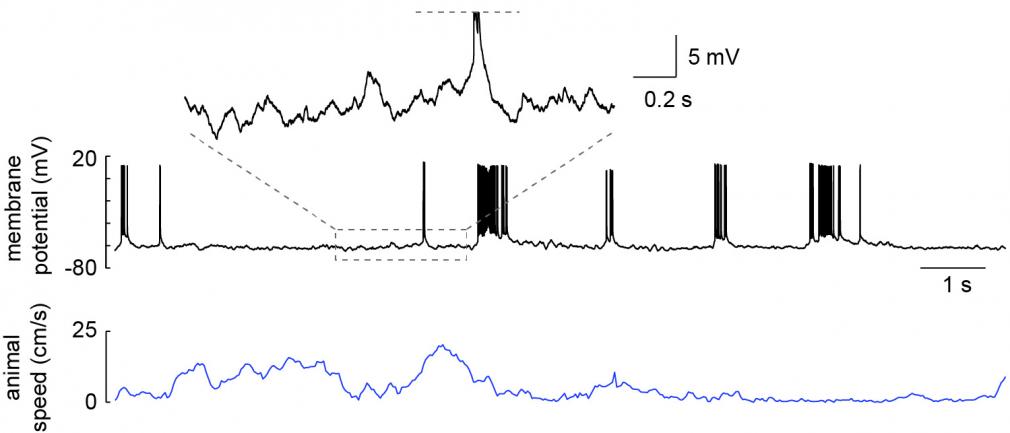
We use multiple techniques to understand spatial processing and memory. We use extracellular recording to monitor the simultaneous activity of large numbers of neurons while an animal explores new spatial environments (Rich et al 2014) and learns locations of importance (e.g. where rewards are). This allows us to get an overall picture of the neural circuits underlying these behaviors. In order to monitor and manipulate the activity of a single neuron in much greater detail, we have developed methods to do intracellular recording in freely moving animals (Lee et al 2014). This allows us to more directly study the cellular mechanisms of spatial processing during behavior (Epsztein et al 2011, Lee et al 2012). We also employ methods to monitor and manipulate activity in animals fixed in place while behaving in virtual spaces, which allows precise control of the sensory environment that the animals interact with.
Dr. Lee is a principal investigator in the Department of Neurology at Beth Israel Deaconess Medical Center and an associate professor of neurology at Harvard Medical School. He was a group leader at HHMI’s Janelia Research Campus from 2008-2023.
albertleeneuro.org
48-meter-long track used to study the large-scale statistical structure of place fields across a population of hippocampal neurons. From Rich et. al.2014.
Whole-cell recording of a hippocampal CA1 pyramidal neuron from a freely moving rat. Recording was obtained in an awake head-fixed animal that was then released for unrestrained behavior. From Lee et al. 2014.