Main Menu (Mobile)- Block
- Overview
-
Support Teams
- Overview
- Anatomy and Histology
- Cryo-Electron Microscopy
- Electron Microscopy
- Flow Cytometry
- Gene Targeting and Transgenics
- High Performance Computing
- Immortalized Cell Line Culture
- Integrative Imaging
- Invertebrate Shared Resource
- Janelia Experimental Technology
- Mass Spectrometry
- Media Prep
- Molecular Genomics
- Stem Cell & Primary Culture
- Project Pipeline Support
- Project Technical Resources
- Quantitative Genomics
- Scientific Computing
- Viral Tools
- Vivarium
- Open Science
- You + Janelia
- About Us
Main Menu - Block
- Overview
- Anatomy and Histology
- Cryo-Electron Microscopy
- Electron Microscopy
- Flow Cytometry
- Gene Targeting and Transgenics
- High Performance Computing
- Immortalized Cell Line Culture
- Integrative Imaging
- Invertebrate Shared Resource
- Janelia Experimental Technology
- Mass Spectrometry
- Media Prep
- Molecular Genomics
- Stem Cell & Primary Culture
- Project Pipeline Support
- Project Technical Resources
- Quantitative Genomics
- Scientific Computing
- Viral Tools
- Vivarium
The VT GAL4, LexA, and split-GAL4 driver line collections for targeted expression in the Drosophila nervous system
About the Fly Lines
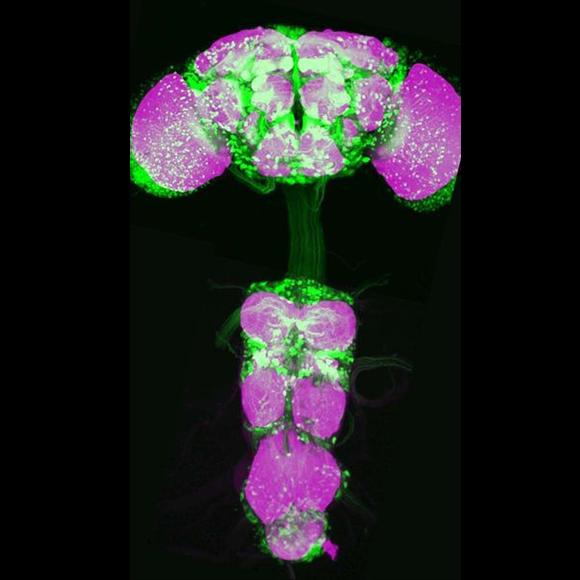
Researchers now at Janelia generated over 18,000 driver lines (the VT collection) that exploit the GAL4, LexA, and split-GAL4 systems to express transgenes in distinct and highly specific cell types in Drosophila. In addition, approximately 1,000 of the most useful VT GAL4 lines are available from the Vienna Drosophila Resource Center.
The GAL4/UAS system is a robust and versatile method for targeted transgene expression in Drosophila, and large collections of GAL4 lines have previously been generated. For example, Janelia researchers generated a total of 8,166 enhancer-GAL4 lines (the “VT” or “Vienna Tiles” lines) containing a predicted ~2kb enhancer region, or in some cases, a promoter region, cloned into the pBPGUw vector or its promoterless equivalent pBPGw. These GAL4 driver constructs were inserted by site-specific recombination into the attP2 landing site on chromosome arm 3L.
Details on the expression pattern, incorporation with the LexA/lexAop system, and use in intersectional targeting may be found in the BioArXiv publication, Tirion et al. 2017.
Our data set of 41,492 confocal images of male brains from 14,635 lines can be accessed online at Virtual Fly Brain. Many of the VT GAL4 and LexA lines have also been independently imaged in female brains and ventral nerve cords at Janelia (https://flweb.janelia.org/cgi-bin/flew.cgi). Almost all of the splitGAL4 hemidriver lines are available from the Bloomington Drosophila Stock Center.