A new imaging method follows young neurons in a developing embryo as they progress from a messy jumble of cells into a coordinated control center. The approach lets scientists track development and emerging cell function simultaneously across entire circuits.
Neurons, take your places.
The cells that eventually become neurons must first travel across the embryo to reach their final destinations in the nervous system. There, they blossom from undefined cells into neurons with specific roles, working together in circuits to direct an animal’s behavior.
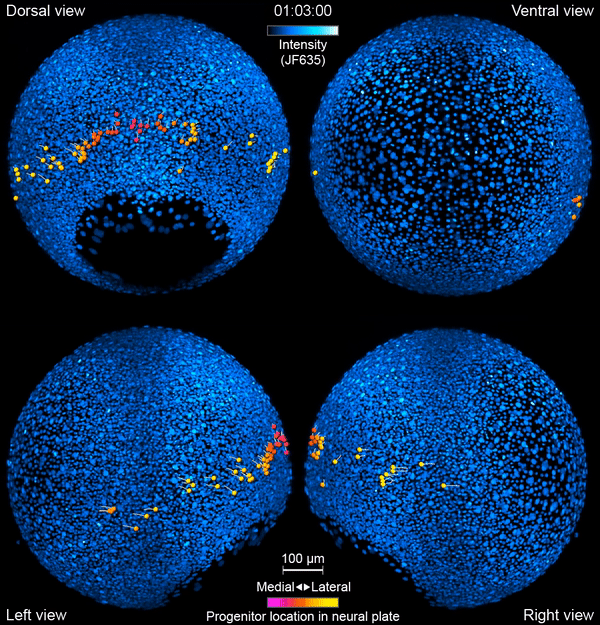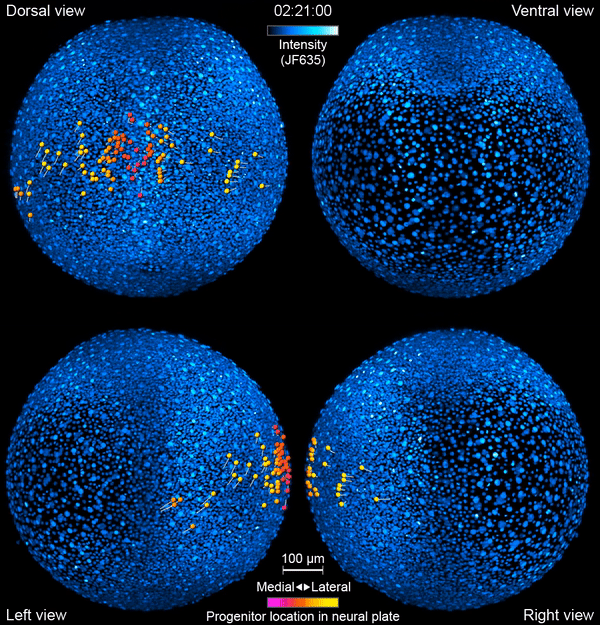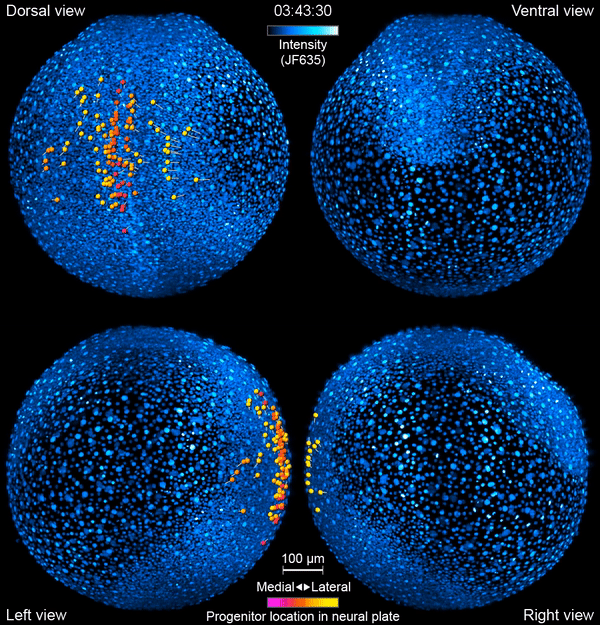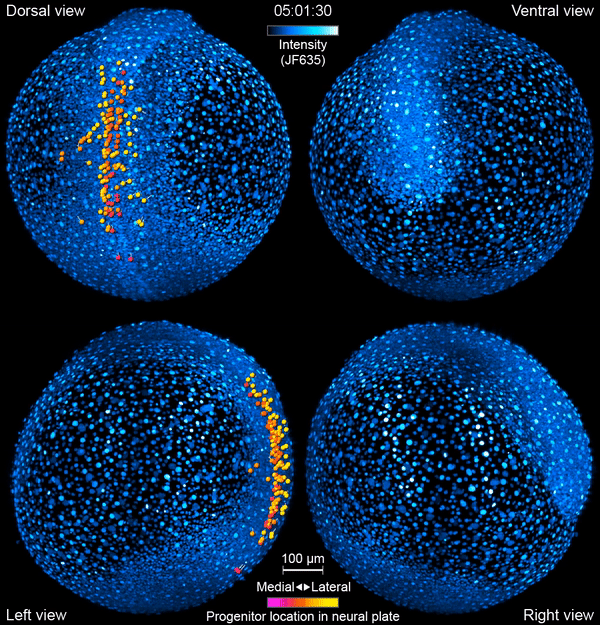
But exactly how this journey unfolds is still somewhat mysterious. “There are lots of things we’re guessing are happening that we don’t actually see,” says Yinan Wan, a researcher at the Howard Hughes Medical Institute’s Janelia Research Campus. Now, Wan and her colleagues have developed tools to watch the action directly – in living animals.
Like a home video series recording a kid from babyhood to their first day of kindergarten, the team’s new videos follow zebrafish neurons from their origin to when they link up into circuits that coordinate movements in the body. The researchers report their work September 26, 2019, in the journal Cell. It’s the first time that scientists have simultaneously tracked the developmental origins, movement, and functional activity of all neurons as they form an entire circuit, from start to finish.
Connecting the physical location and developmental history of young neurons to their emerging roles in the nervous system offers a new window into the way brains organize themselves, says Wan, who is now at Stanford University. “I view these tools as a platform for understanding neural development.”
A new view
It’s taken Wan, Janelia Group Leader Philipp Keller, and the rest of their team some seven years to build the tools needed to collect and analyze data from developing neurons.
“You need technology that allows you to follow, at the single-cell level, the development of an entire embryo,” Keller says. It’s not too hard to find a microscope that can image a large area, or capture the tiniest details, or take pictures really fast. But usually those perks come with tradeoffs. For this experiment, Keller’s team needed a microscope that could do all those things at once, and on a fragile living organism, to boot. “You don’t want to make any compromises – it needs to be perfect,” he says.
Their starting place was a light-sheet microscope that Keller and others at Janelia had developed. Last year, the team used similar technology to watch cells divide, move, and begin to form organs in developing mouse embryos. This time, Keller’s group focused on the nervous system, tracking not just where cells were but also what each was doing.
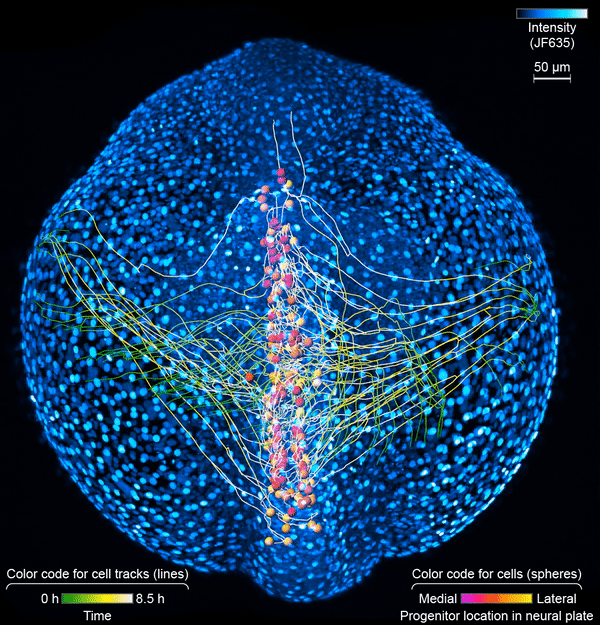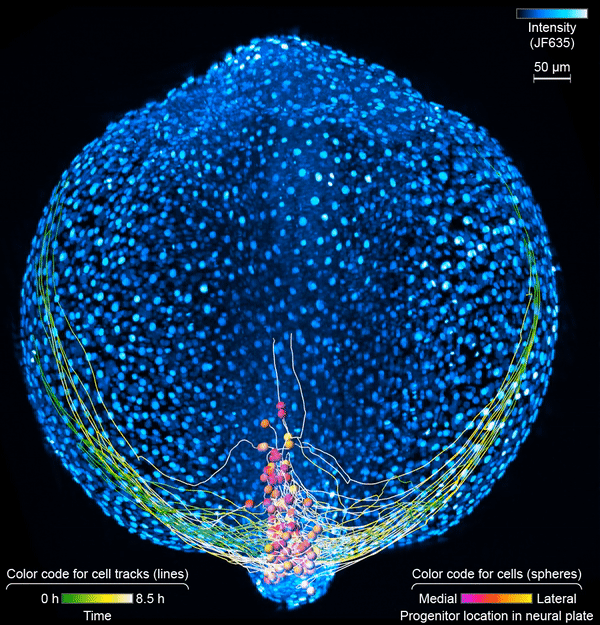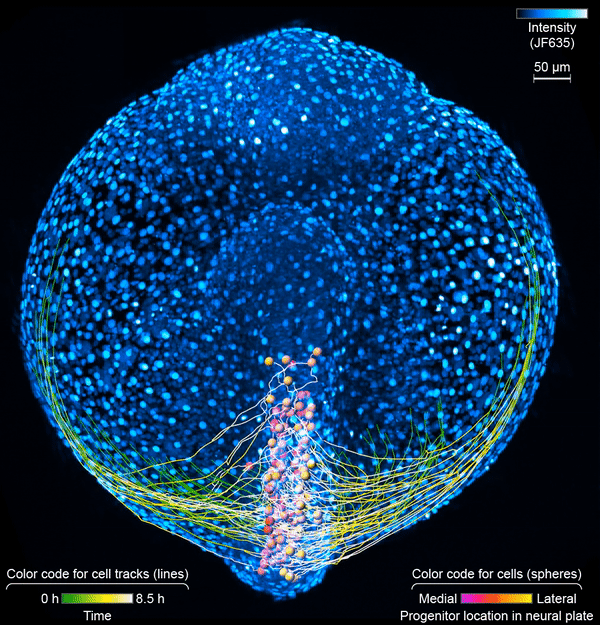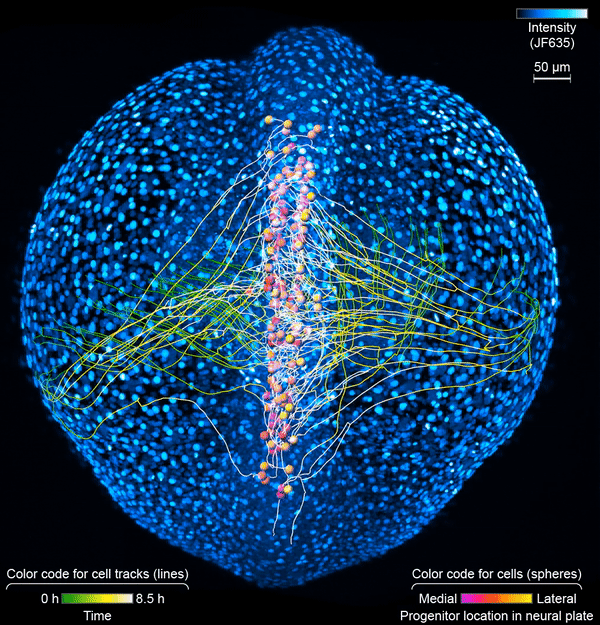
First, the scientists engineered zebrafish to include a small molecule that makes every cell in the fish glow. In the embryo’s neurons, they also tracked a molecule that reports the neurons’ activity, plus a handful of key proteins that are only present when a cell has taken on a specific function – clues to what that cell is actually doing in the body. All together, that information let the team distinguish different kinds of neurons and watch as these cells assumed their roles.
Keller’s group then put the zebrafish embryos under the scope for 14 hours, capturing the movements of all cells and following the cells’ activity at a speed of four 3-D images per second – several million high-resolution snapshots in total. Algorithms developed by Wan and others in the lab helped them reconstruct the paths of individual neurons. And Janelia collaborators Ziqiang Wei and Shaul Druckmann developed computational techniques for analyzing the neurons’ activity patterns.
Over time, the microscope images revealed cells on the move and finding their places, then assuming specific roles and wiring together into circuits. And it showed, at the single-cell level, how highly coordinated network activity first emerges and gives rise to the zebrafish’s earliest behaviors.
“A lot of computational neuroscience right now revolves around how to understand patterns of activity in populations of neurons,” says Druckmann, who is now based at Stanford University. “Developmental studies like these add a whole new dimension: making sense not just of the current population dynamics, but how these patterns develop and change over time.”
The origins of movement
The circuit that the team homed in on – the motor circuit in the zebrafish spinal cord – is the first to develop in fish, Wan says. It’s been extensively studied from many angles. But when it comes to understanding how cells in the circuit mature and begin to work together, “there’s just a big knowledge gap,” she says. The team’s work starts to explain how coordinated movement emerges from disorder.
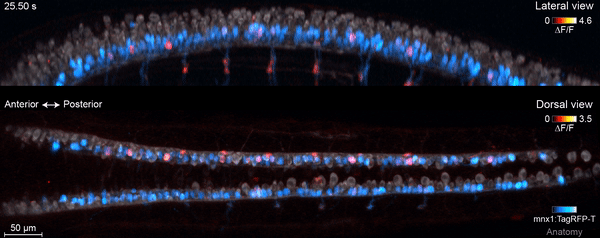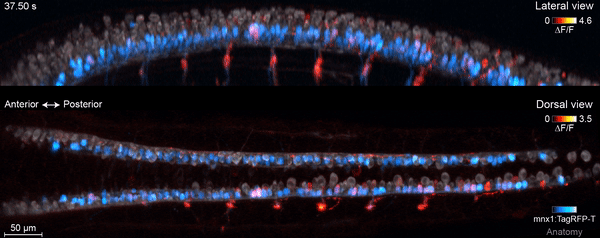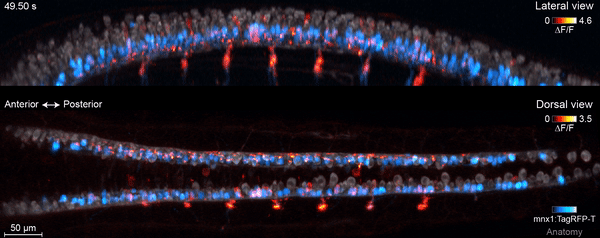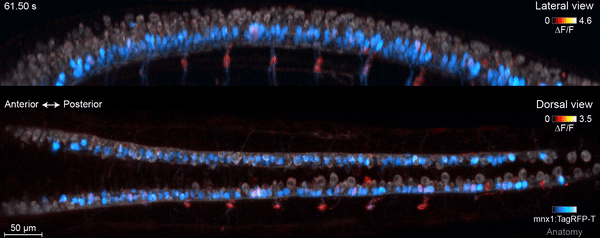
The motor circuit has both motor neurons, which talk to muscles, and interneurons, which bring in signals from other neurons and sometimes act as pacemakers. As the circuit took shape in the developing fish, motor neurons were the first cells to start sending messages, the team found. That was a surprise, Keller says. Scientists had thought motor neurons would likely follow the lead of other cells in this process.
“We have reconstructed the development of individual organs and even entire embryos in the past,” Keller says, “but we have never before combined this with system-wide, high-speed functional imaging of the same cells.” Looking at brain cell development and function together lets the researchers piece together how neural function emerges – at the single-cell level.
###
Citation
Yinan Wan, Ziqiang Wei, Loren L. Looger, Minoru Koyama, Shaul Druckmann, and Philipp J. Keller. “Single-cell reconstruction of emerging population activity in an entire developing circuit.” Cell. Published online September 26, 2019. doi: 10.1016/j.cell.2019.08.039
Media Contacts
Laurel Hamers
hamersl@janelia.hhmi.org





