Filter
Associated Lab
- Aguilera Castrejon Lab (17) Apply Aguilera Castrejon Lab filter
- Ahrens Lab (68) Apply Ahrens Lab filter
- Aso Lab (42) Apply Aso Lab filter
- Baker Lab (38) Apply Baker Lab filter
- Betzig Lab (115) Apply Betzig Lab filter
- Beyene Lab (14) Apply Beyene Lab filter
- Bock Lab (17) Apply Bock Lab filter
- Branson Lab (54) Apply Branson Lab filter
- Card Lab (43) Apply Card Lab filter
- Cardona Lab (64) Apply Cardona Lab filter
- Chklovskii Lab (13) Apply Chklovskii Lab filter
- Clapham Lab (15) Apply Clapham Lab filter
- Cui Lab (19) Apply Cui Lab filter
- Darshan Lab (12) Apply Darshan Lab filter
- Dennis Lab (1) Apply Dennis Lab filter
- Dickson Lab (46) Apply Dickson Lab filter
- Druckmann Lab (25) Apply Druckmann Lab filter
- Dudman Lab (52) Apply Dudman Lab filter
- Eddy/Rivas Lab (30) Apply Eddy/Rivas Lab filter
- Egnor Lab (11) Apply Egnor Lab filter
- Espinosa Medina Lab (21) Apply Espinosa Medina Lab filter
- Feliciano Lab (10) Apply Feliciano Lab filter
- Fetter Lab (41) Apply Fetter Lab filter
- FIB-SEM Technology (1) Apply FIB-SEM Technology filter
- Fitzgerald Lab (29) Apply Fitzgerald Lab filter
- Freeman Lab (15) Apply Freeman Lab filter
- Funke Lab (42) Apply Funke Lab filter
- Gonen Lab (91) Apply Gonen Lab filter
- Grigorieff Lab (62) Apply Grigorieff Lab filter
- Harris Lab (64) Apply Harris Lab filter
- Heberlein Lab (94) Apply Heberlein Lab filter
- Hermundstad Lab (29) Apply Hermundstad Lab filter
- Hess Lab (79) Apply Hess Lab filter
- Ilanges Lab (2) Apply Ilanges Lab filter
- Jayaraman Lab (47) Apply Jayaraman Lab filter
- Ji Lab (33) Apply Ji Lab filter
- Johnson Lab (6) Apply Johnson Lab filter
- Kainmueller Lab (19) Apply Kainmueller Lab filter
- Karpova Lab (14) Apply Karpova Lab filter
- Keleman Lab (13) Apply Keleman Lab filter
- Keller Lab (76) Apply Keller Lab filter
- Koay Lab (18) Apply Koay Lab filter
- Lavis Lab (154) Apply Lavis Lab filter
- Lee (Albert) Lab (34) Apply Lee (Albert) Lab filter
- Leonardo Lab (23) Apply Leonardo Lab filter
- Li Lab (30) Apply Li Lab filter
- Lippincott-Schwartz Lab (178) Apply Lippincott-Schwartz Lab filter
- Liu (Yin) Lab (7) Apply Liu (Yin) Lab filter
- Liu (Zhe) Lab (64) Apply Liu (Zhe) Lab filter
- Looger Lab (138) Apply Looger Lab filter
- Magee Lab (49) Apply Magee Lab filter
- Menon Lab (18) Apply Menon Lab filter
- Murphy Lab (13) Apply Murphy Lab filter
- O'Shea Lab (7) Apply O'Shea Lab filter
- Otopalik Lab (13) Apply Otopalik Lab filter
- Pachitariu Lab (49) Apply Pachitariu Lab filter
- Pastalkova Lab (18) Apply Pastalkova Lab filter
- Pavlopoulos Lab (19) Apply Pavlopoulos Lab filter
- Pedram Lab (15) Apply Pedram Lab filter
- Podgorski Lab (16) Apply Podgorski Lab filter
- Reiser Lab (53) Apply Reiser Lab filter
- Riddiford Lab (44) Apply Riddiford Lab filter
- Romani Lab (49) Apply Romani Lab filter
- Rubin Lab (147) Apply Rubin Lab filter
- Saalfeld Lab (64) Apply Saalfeld Lab filter
- Satou Lab (16) Apply Satou Lab filter
- Scheffer Lab (38) Apply Scheffer Lab filter
- Schreiter Lab (68) Apply Schreiter Lab filter
- Sgro Lab (21) Apply Sgro Lab filter
- Shroff Lab (31) Apply Shroff Lab filter
- Simpson Lab (23) Apply Simpson Lab filter
- Singer Lab (80) Apply Singer Lab filter
- Spruston Lab (97) Apply Spruston Lab filter
- Stern Lab (158) Apply Stern Lab filter
- Sternson Lab (54) Apply Sternson Lab filter
- Stringer Lab (39) Apply Stringer Lab filter
- Svoboda Lab (135) Apply Svoboda Lab filter
- Tebo Lab (35) Apply Tebo Lab filter
- Tervo Lab (9) Apply Tervo Lab filter
- Tillberg Lab (21) Apply Tillberg Lab filter
- Tjian Lab (64) Apply Tjian Lab filter
- Truman Lab (88) Apply Truman Lab filter
- Turaga Lab (53) Apply Turaga Lab filter
- Turner Lab (39) Apply Turner Lab filter
- Vale Lab (8) Apply Vale Lab filter
- Voigts Lab (3) Apply Voigts Lab filter
- Wang (Meng) Lab (27) Apply Wang (Meng) Lab filter
- Wang (Shaohe) Lab (25) Apply Wang (Shaohe) Lab filter
- Wu Lab (9) Apply Wu Lab filter
- Zlatic Lab (28) Apply Zlatic Lab filter
- Zuker Lab (25) Apply Zuker Lab filter
Associated Project Team
- CellMap (12) Apply CellMap filter
- COSEM (3) Apply COSEM filter
- FIB-SEM Technology (5) Apply FIB-SEM Technology filter
- Fly Descending Interneuron (12) Apply Fly Descending Interneuron filter
- Fly Functional Connectome (14) Apply Fly Functional Connectome filter
- Fly Olympiad (5) Apply Fly Olympiad filter
- FlyEM (56) Apply FlyEM filter
- FlyLight (50) Apply FlyLight filter
- GENIE (47) Apply GENIE filter
- Integrative Imaging (7) Apply Integrative Imaging filter
- Larval Olympiad (2) Apply Larval Olympiad filter
- MouseLight (18) Apply MouseLight filter
- NeuroSeq (1) Apply NeuroSeq filter
- ThalamoSeq (1) Apply ThalamoSeq filter
- Tool Translation Team (T3) (28) Apply Tool Translation Team (T3) filter
- Transcription Imaging (49) Apply Transcription Imaging filter
Publication Date
- 2025 (219) Apply 2025 filter
- 2024 (212) Apply 2024 filter
- 2023 (158) Apply 2023 filter
- 2022 (192) Apply 2022 filter
- 2021 (194) Apply 2021 filter
- 2020 (196) Apply 2020 filter
- 2019 (202) Apply 2019 filter
- 2018 (232) Apply 2018 filter
- 2017 (217) Apply 2017 filter
- 2016 (209) Apply 2016 filter
- 2015 (252) Apply 2015 filter
- 2014 (236) Apply 2014 filter
- 2013 (194) Apply 2013 filter
- 2012 (190) Apply 2012 filter
- 2011 (190) Apply 2011 filter
- 2010 (161) Apply 2010 filter
- 2009 (158) Apply 2009 filter
- 2008 (140) Apply 2008 filter
- 2007 (106) Apply 2007 filter
- 2006 (92) Apply 2006 filter
- 2005 (67) Apply 2005 filter
- 2004 (57) Apply 2004 filter
- 2003 (58) Apply 2003 filter
- 2002 (39) Apply 2002 filter
- 2001 (28) Apply 2001 filter
- 2000 (29) Apply 2000 filter
- 1999 (14) Apply 1999 filter
- 1998 (18) Apply 1998 filter
- 1997 (16) Apply 1997 filter
- 1996 (10) Apply 1996 filter
- 1995 (18) Apply 1995 filter
- 1994 (12) Apply 1994 filter
- 1993 (10) Apply 1993 filter
- 1992 (6) Apply 1992 filter
- 1991 (11) Apply 1991 filter
- 1990 (11) Apply 1990 filter
- 1989 (6) Apply 1989 filter
- 1988 (1) Apply 1988 filter
- 1987 (7) Apply 1987 filter
- 1986 (4) Apply 1986 filter
- 1985 (5) Apply 1985 filter
- 1984 (2) Apply 1984 filter
- 1983 (2) Apply 1983 filter
- 1982 (3) Apply 1982 filter
- 1981 (3) Apply 1981 filter
- 1980 (1) Apply 1980 filter
- 1979 (1) Apply 1979 filter
- 1976 (2) Apply 1976 filter
- 1973 (1) Apply 1973 filter
- 1970 (1) Apply 1970 filter
- 1967 (1) Apply 1967 filter
Type of Publication
4194 Publications
Showing 3371-3380 of 4194 resultsMitochondria are essential organelles whose biogenesis, structure, and function are regulated by many signaling pathways. In this study we present evidence that, in hippocampal neurons, activation of the Sonic hedgehog (Shh) signaling pathway impacts multiple aspects of mitochondria. Mitochondrial mass was increased significantly in neurons treated with Shh. Using biochemical and fluorescence imaging analyses, we show that Shh signaling activity reduces mitochondrial fission and promotes mitochondrial elongation, at least in part, via suppression of the mitochondrial fission protein dynamin-like GTPase Drp1. Mitochondria from Shh-treated neurons were more electron-dense as revealed by electron microscopy, and had higher membrane potential and respiratory activity. We further show that Shh protects neurons against a variety of stresses, including the mitochondrial poison rotenone, amyloid β-peptide, hydrogen peroxide, and high levels of glutamate. Collectively, our data suggest a link between Shh pathway activity and the physiological properties of mitochondria in hippocampal neurons.
How does the mammalian retina detect motion? This classic problem in visual neuroscience has remained unsolved for 50 years. In search of clues, here we reconstruct Off-type starburst amacrine cells (SACs) and bipolar cells (BCs) in serial electron microscopic images with help from EyeWire, an online community of 'citizen neuroscientists'. On the basis of quantitative analyses of contact area and branch depth in the retina, we find evidence that one BC type prefers to wire with a SAC dendrite near the SAC soma, whereas another BC type prefers to wire far from the soma. The near type is known to lag the far type in time of visual response. A mathematical model shows how such 'space-time wiring specificity' could endow SAC dendrites with receptive fields that are oriented in space-time and therefore respond selectively to stimuli that move in the outward direction from the soma.
In the development of multicellular organisms a diversity of cell types differentiate at specific positions. Spacing patterns, in which an array of two or more cell types forms from a uniform field of cells, are a common feature of development. Identical precursor cells may adopt different fates because of competition and inhibition between them. Such a pattern in the developing Drosophila eye is the evenly spaced array of R8 cells, around which other cell types are subsequently recruited. Genetic studies suggest that the scabrous mutation disrupts a signal produced by R8 cells that inhibits other cells from also becoming R8 cells. The scabrous locus was cloned, and it appears to encode a secreted protein partly related to the beta and gamma chains of fibrinogen. It is proposed that the sca locus encodes a lateral inhibitor of R8 differentiation. The roles of the Drosophila EGF-receptor homologue (DER) and Notch genes in this process were also investigated.
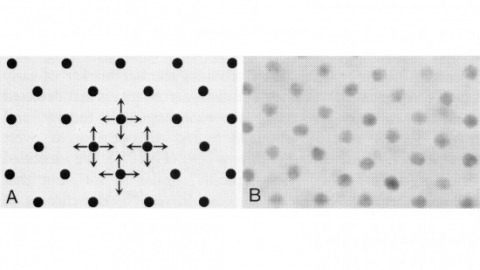
A method is described that yields a series of (D+1)-element wave-vector sets giving rise to (D=2 or 3)-dimensional coherent sparse lattices of any desired Bravais symmetry and primitive cell shape, but of increasing period relative to the excitation wavelength. By applying lattice symmetry operations to any of these sets, composite lattices of N>D+1 waves are constructed, having increased spatial frequency content but unchanged crystal group symmetry and periodicity. Optical lattices of widely spaced excitation maxima of diffraction-limited confinement and controllable polarization can thereby be created, possibly useful for quan- tum optics, lithography, or multifocal microscopy.
Commentary: Develops a formalism to find a set of wavevectors that create a periodic optical lattice of any desired Bravais symmetry by the mutual interference of the corresponding plane waves. Discovers two new classes of optical lattices, sparse and composite, that together permit the creation of widely spaced, tightly confined excitation maxima in 3D potentially suitable for high speed volumetric live cell imaging. The implementation of this idea was derailed by our exclusive focus on PALM at the time, and many of its goals have since been reached with our Bessel beam plane illumination microscope. Nevertheless, sparse and composite optical lattices may prove useful in atomic physics or for the fabrication of 3D nanostructures.
Mitral/tufted cells of the olfactory bulb receive odorant information from receptor neurons and transmit this information to the cortex. Studies in awake behaving animals have found that sustained responses of mitral cells to odorants are rare, suggesting sparse combinatorial representation of the odorants. Careful alignment of mitral cell firing with the phase of the respiration cycle revealed brief transient activity in the larger population of mitral cells, which respond to odorants during a small fraction of the respiration cycle. Responses of these cells are therefore temporally sparse. Here, we propose a mathematical model for the olfactory bulb network that can reproduce both combinatorially and temporally sparse mitral cell codes. We argue that sparse codes emerge as a result of the balance between mitral cells’ excitatory inputs and inhibition provided by the granule cells. Our model suggests functional significance for the dendrodendritic synapses mediating interactions between mitral and granule cells.
We propose a version of least-mean-square (LMS) algorithm for sparse system identification. Our algorithm called online linearized Bregman iteration (OLBI) is derived from minimizing the cumulative prediction error squared along with an l 1 -l 2 norm regularizer. By systematically treating the non-differentiable regularizer we arrive at a simple two-step iteration. We demonstrate that OLBI is bias free and compare its operation with existing sparse LMS algorithms by rederiving them in the online convex optimization framework. We perform convergence analysis of OLBI for white input signals and derive theoretical expressions for the steady state mean square deviations (MSD). We demonstrate numerically that OLBI improves the performance of LMS type algorithms for signals generated from sparse tap weights.
Responses of mitral cells represent the results of the first stage of odor processing in the olfactory bulb. Most of our knowledge about mitral cell activity has been obtained from recordings in anesthetized animals. We compared odor-elicited changes in firing rate of mitral cells in awake behaving mice and in anesthetized mice. We show that odor-elicited changes in mitral cell firing rate were larger and more frequently observed in the anesthetized than in the awake condition. Only 27% of mitral cells that showed a response to odors in the anesthetized state were also odor responsive in the awake state. The amplitude of their response in the awake state was smaller, and some of the responses changed sign compared with their responses in the anesthetized state. The odor representation in the olfactory bulb is therefore sparser in awake behaving mice than in anesthetized preparations. A qualitative explanation of the mechanism responsible for this phenomenon is proposed.
Electrical microstimulation can establish causal links between the activity of groups of neurons and perceptual and cognitive functions. However, the number and identities of neurons microstimulated, as well as the number of action potentials evoked, are difficult to ascertain. To address these issues we introduced the light-gated algal channel channelrhodopsin-2 (ChR2) specifically into a small fraction of layer 2/3 neurons of the mouse primary somatosensory cortex. ChR2 photostimulation in vivo reliably generated stimulus-locked action potentials at frequencies up to 50 Hz. Here we show that naive mice readily learned to detect brief trains of action potentials (five light pulses, 1 ms, 20 Hz). After training, mice could detect a photostimulus firing a single action potential in approximately 300 neurons. Even fewer neurons (approximately 60) were required for longer stimuli (five action potentials, 250 ms). Our results show that perceptual decisions and learning can be driven by extremely brief epochs of cortical activity in a sparse subset of supragranular cortical pyramidal neurons.
Sparse coding may be a general strategy of neural systems for augmenting memory capacity. In Drosophila melanogaster, sparse odor coding by the Kenyon cells of the mushroom body is thought to generate a large number of precisely addressable locations for the storage of odor-specific memories. However, it remains untested how sparse coding relates to behavioral performance. Here we demonstrate that sparseness is controlled by a negative feedback circuit between Kenyon cells and the GABAergic anterior paired lateral (APL) neuron. Systematic activation and blockade of each leg of this feedback circuit showed that Kenyon cells activated APL and APL inhibited Kenyon cells. Disrupting the Kenyon cell-APL feedback loop decreased the sparseness of Kenyon cell odor responses, increased inter-odor correlations and prevented flies from learning to discriminate similar, but not dissimilar, odors. These results suggest that feedback inhibition suppresses Kenyon cell activity to maintain sparse, decorrelated odor coding and thus the odor specificity of memories.
Lipid droplets (LDs) are neutral lipid storage organelles that transfer lipids to various organelles including peroxisomes. Here, we show that the hereditary spastic paraplegia protein M1 Spastin, a membrane-bound AAA ATPase found on LDs, coordinates fatty acid (FA) trafficking from LDs to peroxisomes through two inter-related mechanisms. First, M1 Spastin forms a tethering complex with peroxisomal ABCD1 to promote LD-peroxisome contact formation. Second, M1 Spastin recruits the membrane-shaping ESCRT-III proteins IST1 and CHMP1B to LDs via its MIT domain to facilitate LD-to-peroxisome FA trafficking, possibly through IST1 and CHMP1B modifying LD membrane morphology. Furthermore, M1 Spastin, IST1 and CHMP1B are all required to relieve LDs of lipid peroxidation. The roles of M1 Spastin in tethering LDs to peroxisomes and in recruiting ESCRT-III components to LD-peroxisome contact sites for FA trafficking may help explain the pathogenesis of diseases associated with defective FA metabolism in LDs and peroxisomes.