Filter
Associated Lab
- Aguilera Castrejon Lab (17) Apply Aguilera Castrejon Lab filter
- Ahrens Lab (69) Apply Ahrens Lab filter
- Aso Lab (42) Apply Aso Lab filter
- Baker Lab (38) Apply Baker Lab filter
- Betzig Lab (115) Apply Betzig Lab filter
- Beyene Lab (14) Apply Beyene Lab filter
- Bock Lab (17) Apply Bock Lab filter
- Branson Lab (54) Apply Branson Lab filter
- Card Lab (43) Apply Card Lab filter
- Cardona Lab (64) Apply Cardona Lab filter
- Chklovskii Lab (13) Apply Chklovskii Lab filter
- Clapham Lab (15) Apply Clapham Lab filter
- Cui Lab (19) Apply Cui Lab filter
- Darshan Lab (12) Apply Darshan Lab filter
- Dennis Lab (2) Apply Dennis Lab filter
- Dickson Lab (46) Apply Dickson Lab filter
- Druckmann Lab (25) Apply Druckmann Lab filter
- Dudman Lab (53) Apply Dudman Lab filter
- Eddy/Rivas Lab (30) Apply Eddy/Rivas Lab filter
- Egnor Lab (11) Apply Egnor Lab filter
- Espinosa Medina Lab (21) Apply Espinosa Medina Lab filter
- Feliciano Lab (10) Apply Feliciano Lab filter
- Fetter Lab (41) Apply Fetter Lab filter
- FIB-SEM Technology (1) Apply FIB-SEM Technology filter
- Fitzgerald Lab (29) Apply Fitzgerald Lab filter
- Freeman Lab (15) Apply Freeman Lab filter
- Funke Lab (42) Apply Funke Lab filter
- Gonen Lab (91) Apply Gonen Lab filter
- Grigorieff Lab (62) Apply Grigorieff Lab filter
- Harris Lab (64) Apply Harris Lab filter
- Heberlein Lab (94) Apply Heberlein Lab filter
- Hermundstad Lab (30) Apply Hermundstad Lab filter
- Hess Lab (79) Apply Hess Lab filter
- Ilanges Lab (3) Apply Ilanges Lab filter
- Jayaraman Lab (48) Apply Jayaraman Lab filter
- Ji Lab (33) Apply Ji Lab filter
- Johnson Lab (6) Apply Johnson Lab filter
- Kainmueller Lab (19) Apply Kainmueller Lab filter
- Karpova Lab (14) Apply Karpova Lab filter
- Keleman Lab (13) Apply Keleman Lab filter
- Keller Lab (76) Apply Keller Lab filter
- Koay Lab (18) Apply Koay Lab filter
- Lavis Lab (154) Apply Lavis Lab filter
- Lee (Albert) Lab (34) Apply Lee (Albert) Lab filter
- Leonardo Lab (23) Apply Leonardo Lab filter
- Li Lab (30) Apply Li Lab filter
- Lippincott-Schwartz Lab (178) Apply Lippincott-Schwartz Lab filter
- Liu (Yin) Lab (7) Apply Liu (Yin) Lab filter
- Liu (Zhe) Lab (64) Apply Liu (Zhe) Lab filter
- Looger Lab (138) Apply Looger Lab filter
- Magee Lab (49) Apply Magee Lab filter
- Menon Lab (18) Apply Menon Lab filter
- Murphy Lab (13) Apply Murphy Lab filter
- O'Shea Lab (7) Apply O'Shea Lab filter
- Otopalik Lab (13) Apply Otopalik Lab filter
- Pachitariu Lab (49) Apply Pachitariu Lab filter
- Pastalkova Lab (18) Apply Pastalkova Lab filter
- Pavlopoulos Lab (19) Apply Pavlopoulos Lab filter
- Pedram Lab (15) Apply Pedram Lab filter
- Podgorski Lab (16) Apply Podgorski Lab filter
- Reiser Lab (54) Apply Reiser Lab filter
- Riddiford Lab (44) Apply Riddiford Lab filter
- Romani Lab (49) Apply Romani Lab filter
- Rubin Lab (148) Apply Rubin Lab filter
- Saalfeld Lab (64) Apply Saalfeld Lab filter
- Satou Lab (16) Apply Satou Lab filter
- Scheffer Lab (38) Apply Scheffer Lab filter
- Schreiter Lab (69) Apply Schreiter Lab filter
- Sgro Lab (21) Apply Sgro Lab filter
- Shroff Lab (31) Apply Shroff Lab filter
- Simpson Lab (23) Apply Simpson Lab filter
- Singer Lab (80) Apply Singer Lab filter
- Spruston Lab (97) Apply Spruston Lab filter
- Stern Lab (158) Apply Stern Lab filter
- Sternson Lab (54) Apply Sternson Lab filter
- Stringer Lab (39) Apply Stringer Lab filter
- Svoboda Lab (135) Apply Svoboda Lab filter
- Tebo Lab (35) Apply Tebo Lab filter
- Tervo Lab (9) Apply Tervo Lab filter
- Tillberg Lab (21) Apply Tillberg Lab filter
- Tjian Lab (64) Apply Tjian Lab filter
- Truman Lab (88) Apply Truman Lab filter
- Turaga Lab (53) Apply Turaga Lab filter
- Turner Lab (39) Apply Turner Lab filter
- Vale Lab (8) Apply Vale Lab filter
- Voigts Lab (4) Apply Voigts Lab filter
- Wang (Meng) Lab (27) Apply Wang (Meng) Lab filter
- Wang (Shaohe) Lab (25) Apply Wang (Shaohe) Lab filter
- Wu Lab (9) Apply Wu Lab filter
- Zlatic Lab (28) Apply Zlatic Lab filter
- Zuker Lab (25) Apply Zuker Lab filter
Associated Project Team
- CellMap (12) Apply CellMap filter
- COSEM (3) Apply COSEM filter
- FIB-SEM Technology (5) Apply FIB-SEM Technology filter
- Fly Descending Interneuron (12) Apply Fly Descending Interneuron filter
- Fly Functional Connectome (14) Apply Fly Functional Connectome filter
- Fly Olympiad (5) Apply Fly Olympiad filter
- FlyEM (56) Apply FlyEM filter
- FlyLight (50) Apply FlyLight filter
- GENIE (47) Apply GENIE filter
- Integrative Imaging (7) Apply Integrative Imaging filter
- Larval Olympiad (2) Apply Larval Olympiad filter
- MouseLight (18) Apply MouseLight filter
- NeuroSeq (1) Apply NeuroSeq filter
- ThalamoSeq (1) Apply ThalamoSeq filter
- Tool Translation Team (T3) (28) Apply Tool Translation Team (T3) filter
- Transcription Imaging (49) Apply Transcription Imaging filter
Publication Date
- 2025 (227) Apply 2025 filter
- 2024 (212) Apply 2024 filter
- 2023 (158) Apply 2023 filter
- 2022 (192) Apply 2022 filter
- 2021 (194) Apply 2021 filter
- 2020 (196) Apply 2020 filter
- 2019 (202) Apply 2019 filter
- 2018 (232) Apply 2018 filter
- 2017 (217) Apply 2017 filter
- 2016 (209) Apply 2016 filter
- 2015 (252) Apply 2015 filter
- 2014 (236) Apply 2014 filter
- 2013 (194) Apply 2013 filter
- 2012 (190) Apply 2012 filter
- 2011 (190) Apply 2011 filter
- 2010 (161) Apply 2010 filter
- 2009 (158) Apply 2009 filter
- 2008 (140) Apply 2008 filter
- 2007 (106) Apply 2007 filter
- 2006 (92) Apply 2006 filter
- 2005 (67) Apply 2005 filter
- 2004 (57) Apply 2004 filter
- 2003 (58) Apply 2003 filter
- 2002 (39) Apply 2002 filter
- 2001 (28) Apply 2001 filter
- 2000 (29) Apply 2000 filter
- 1999 (14) Apply 1999 filter
- 1998 (18) Apply 1998 filter
- 1997 (16) Apply 1997 filter
- 1996 (10) Apply 1996 filter
- 1995 (18) Apply 1995 filter
- 1994 (12) Apply 1994 filter
- 1993 (10) Apply 1993 filter
- 1992 (6) Apply 1992 filter
- 1991 (11) Apply 1991 filter
- 1990 (11) Apply 1990 filter
- 1989 (6) Apply 1989 filter
- 1988 (1) Apply 1988 filter
- 1987 (7) Apply 1987 filter
- 1986 (4) Apply 1986 filter
- 1985 (5) Apply 1985 filter
- 1984 (2) Apply 1984 filter
- 1983 (2) Apply 1983 filter
- 1982 (3) Apply 1982 filter
- 1981 (3) Apply 1981 filter
- 1980 (1) Apply 1980 filter
- 1979 (1) Apply 1979 filter
- 1976 (2) Apply 1976 filter
- 1973 (1) Apply 1973 filter
- 1970 (1) Apply 1970 filter
- 1967 (1) Apply 1967 filter
Type of Publication
4202 Publications
Showing 3781-3790 of 4202 resultsMucin domains are densely O-glycosylated modular protein domains that are found in a wide variety of cell surface and secreted proteins. Mucin-domain glycoproteins are known to be key players in a host of human diseases, especially cancer, wherein mucin expression and glycosylation patterns are altered. Mucin biology has been difficult to study at the molecular level, in part, because methods to manipulate and structurally characterize mucin domains are lacking. Here, we demonstrate that secreted protease of C1 esterase inhibitor (StcE), a bacterial protease from Escherichia coli, cleaves mucin domains by recognizing a discrete peptide- and glycan-based motif. We exploited StcE's unique properties to improve sequence coverage, glycosite mapping, and glycoform analysis of recombinant human mucins by mass spectrometry. We also found that StcE digests cancer-associated mucins from cultured cells and from ascites fluid derived from patients with ovarian cancer. Finally, using StcE, we discovered that sialic acid-binding Ig-type lectin-7 (Siglec-7), a glycoimmune checkpoint receptor, selectively binds sialomucins as biological ligands, whereas the related receptor Siglec-9 does not. Mucin-selective proteolysis, as exemplified by StcE, is therefore a powerful tool for the study of mucin domain structure and function.
Connectomics has focused primarily on the mapping of synaptic links in the brain; yet it is well established that extrasynaptic volume transmission, especially via monoamines and neuropeptides, is also critical to brain function and occurs primarily outside the synaptic connectome. We have mapped the putative monoamine connections, as well as a subset of neuropeptide connections, in C. elegans based on new and published gene expression data. The monoamine and neuropeptide networks exhibit distinct topological properties, with the monoamine network displaying a highly disassortative star-like structure with a rich-club of interconnected broadcasting hubs, and the neuropeptide network showing a more recurrent, highly clustered topology. Despite the low degree of overlap between the extrasynaptic (or wireless) and synaptic (or wired) connectomes, we find highly significant multilink motifs of interaction, pinpointing locations in the network where aminergic and neuropeptide signalling modulate synaptic activity. Thus, the C. elegans connectome can be mapped as a multiplex network with synaptic, gap junction, and neuromodulator layers representing alternative modes of interaction between neurons. This provides a new topological plan for understanding how aminergic and peptidergic modulation of behaviour is achieved by specific motifs and loci of integration between hard-wired synaptic or junctional circuits and extrasynaptic signals wirelessly broadcast from a small number of modulatory neurons.
B cell activation is initiated by the binding of antigen to the B cell receptor (BCR). Here we used dSTORM super resolution imaging to characterize the nanoscale spatial organization of IgM and IgG BCRs on the surfaces of resting and antigen-activated human peripheral blood B cells. We provide insights into both the fundamental process of antigen-driven BCR clustering as well as differences in the spatial organization of IgM and IgG BCRs that may contribute to the characteristic differences in the responses of naïve and memory B cells to antigen. We provide evidence that although both IgM and IgG BCRs reside in highly heterogeneous protein islands that vary in both size and number of BCR single molecule localizations, both resting and activated B cells intrinsically maintain a high frequency of single isolated BCR localizations, which likely represent BCR monomers. IgG BCRs are more clustered than IgM BCRs on resting cells and form larger protein islands following antigen activation. Small dense BCR clusters likely formed via protein-protein interactions are present on the surface of resting cells and antigen activation induces these to come together to form less dense, larger islands, a process likely governed, at least in part, by protein-lipid interactions.
The conserved Ndc80 complex is an essential microtubule-binding component of the kinetochore. Recent findings suggest that the Ndc80 complex influences microtubule dynamics at kinetochores in vivo. However, it was unclear if the Ndc80 complex mediates these effects directly, or by affecting other factors localized at the kinetochore. Using a reconstituted system in vitro, we show that the human Ndc80 complex directly stabilizes the tips of disassembling microtubules and promotes rescue (the transition from microtubule shortening to growth). In vivo, an N-terminal domain in the Ndc80 complex is phosphorylated by the Aurora B kinase. Mutations that mimic phosphorylation of the Ndc80 complex prevent stable kinetochore-microtubule attachment, and mutations that block phosphorylation damp kinetochore oscillations. We find that the Ndc80 complex with Aurora B phosphomimetic mutations is defective at promoting microtubule rescue, even when robustly coupled to disassembling microtubule tips. This impaired ability to affect dynamics is not simply because of weakened microtubule binding, as an N-terminally truncated complex with similar binding affinity is able to promote rescue. Taken together, these results suggest that in addition to regulating attachment stability, Aurora B controls microtubule dynamics through phosphorylation of the Ndc80 complex.
The Ndc80 complex is a key site of regulated kinetochore-microtubule attachment (a process required for cell division), but the molecular mechanism underlying its function remains unknown. Here we present a subnanometre-resolution cryo-electron microscopy reconstruction of the human Ndc80 complex bound to microtubules, sufficient for precise docking of crystal structures of the component proteins. We find that the Ndc80 complex binds the microtubule with a tubulin monomer repeat, recognizing α- and β-tubulin at both intra- and inter-tubulin dimer interfaces in a manner that is sensitive to tubulin conformation. Furthermore, Ndc80 complexes self-associate along protofilaments through interactions mediated by the amino-terminal tail of the NDC80 protein, which is the site of phospho-regulation by Aurora B kinase. The complex’s mode of interaction with the microtubule and its oligomerization suggest a mechanism by which Aurora B could regulate the stability of load-bearing kinetochore-microtubule attachments.
Animal sounds are produced by patterned vibrations of specific organs, but the neural circuits that drive these vibrations are not well defined in any animal. Here we provide a functional and synaptic map of most of the neurons in the Drosophila male ventral nerve cord (the analog of the vertebrate spinal cord) that drive complex, patterned song during courtship. Male Drosophila vibrate their wings toward females during courtship to produce two distinct song modes – pulse and sine song – with characteristic features that signal species identity and male quality. We identified song-producing neural circuits by optogenetically activating and inhibiting identified cell types in the ventral nerve cord (VNC) and by tracing their patterns of synaptic connectivity in the male VNC connectome. The core song circuit consists of at least eight cell types organized into overlapping circuits, where all neurons are required for pulse song and a subset are required for sine song. The pulse and sine circuits each include a feed-forward pathway from brain descending neurons to wing motor neurons, with extensive reciprocal and feed-back connections. We also identify specific neurons that shape the individual features of each song mode. These results reveal commonalities amongst diverse animals in the neural mechanisms that generate diverse motor patterns from a single set of muscles.
The mammalian taste system is responsible for sensing and responding to the five basic taste qualities: sweet, sour, bitter, salty and umami. Previously, we showed that each taste is detected by dedicated taste receptor cells (TRCs) on the tongue and palate epithelium. To understand how TRCs transmit information to higher neural centres, we examined the tuning properties of large ensembles of neurons in the first neural station of the gustatory system. Here, we generated and characterized a collection of transgenic mice expressing a genetically encoded calcium indicator in central and peripheral neurons, and used a gradient refractive index microendoscope combined with high-resolution two-photon microscopy to image taste responses from ganglion neurons buried deep at the base of the brain. Our results reveal fine selectivity in the taste preference of ganglion neurons; demonstrate a strong match between TRCs in the tongue and the principal neural afferents relaying taste information to the brain; and expose the highly specific transfer of taste information between taste cells and the central nervous system.
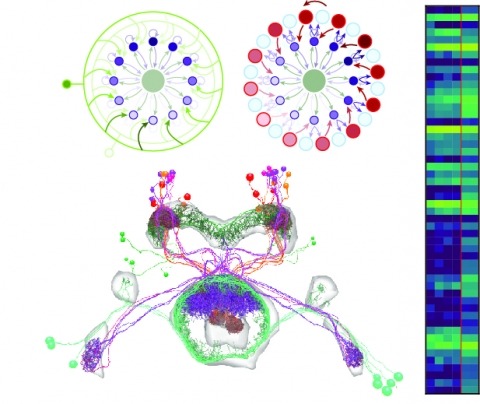
Neural representations of head direction (HD) have been discovered in many species. Theoretical work has proposed that the dynamics associated with these representations are generated, maintained, and updated by recurrent network structures called ring attractors. We evaluated this theorized structure-function relationship by performing electron-microscopy-based circuit reconstruction and RNA profiling of identified cell types in the HD system of Drosophila melanogaster. We identified motifs that have been hypothesized to maintain the HD representation in darkness, update it when the animal turns, and tether it to visual cues. Functional studies provided support for the proposed roles of individual excitatory or inhibitory circuit elements in shaping activity. We also discovered recurrent connections between neuronal arbors with mixed pre- and postsynaptic specializations. Our results confirm that the Drosophila HD network contains the core components of a ring attractor while also revealing unpredicted structural features that might enhance the network's computational power.
The neurophysiology of cells and tissues are monitored electrophysiologically and optically in diverse experiments and species, ranging from flies to humans. Understanding the brain requires integration of data across this diversity, and thus these data must be findable, accessible, interoperable, and reusable (FAIR). This requires a standard language for data and metadata that can coevolve with neuroscience. We describe design and implementation principles for a language for neurophysiology data. Our open-source software (Neurodata Without Borders, NWB) defines and modularizes the interdependent, yet separable, components of a data language. We demonstrate NWB's impact through unified description of neurophysiology data across diverse modalities and species. NWB exists in an ecosystem, which includes data management, analysis, visualization, and archive tools. Thus, the NWB data language enables reproduction, interchange, and reuse of diverse neurophysiology data. More broadly, the design principles of NWB are generally applicable to enhance discovery across biology through data FAIRness.