Hundreds of millions of years ago, single cells joined forces to become multicellular organisms. At the foundation of this multicellular world is the cell surface: the plasma membrane surrounding each cell, where individual units meet and communicate with one another using a language made up of molecules and proteins.
For biologists, being able to decipher the cellular conversation happening at the surface – the proteins that are present, how they interact, and what changes they go through – is key to understanding how an organism functions and, ultimately, how disease happens.
Now, researchers at HHMI’s Janelia Research Campus, Stanford University, and the Broad Institute are developing user-friendly tools to help scientists translate this cellular communication and uncover new details about how cells exchange information.
“If you think about the body as a society of cells, what we really want to do is try to decode the language of that society and how cells communicate with each other,” says Janelia Group Leader Jiefu Li, a senior author on three new publications. “We want to develop methods to figure out that language and use it, step by step, to decode that language.”
The new research not only gives scientists novel insights into cellular communication, but also showcases the potential of proteomics – the study of all the proteins in living cells and organisms – to read the basic language units of the cell and unlock some of biology’s biggest mysteries.
“We started by building tools for proteomics and then used those tools in certain biological questions to make discoveries and also to demonstrate proteomics’ power,” Li says. “We show that proteomics can be applied to many biological questions: from neuroscience to immunology, and from flies to humans.”
Learning the language of cells
Li started developing methods and tools to study proteins at the cell surface while he was a graduate student and postdoc at Stanford. Biologists around the world began using these methods, but many of these researchers contacted Li and his colleagues with the same problem: without a background in chemical biology or proteomics, they were having trouble making sense of the data they were generating.
When Li came to Janelia as a group leader in 2022, he set out to fix the problem. He collaborated with the Scientific Computing Software team to develop PEELing, a user-friendly platform that allows researchers worldwide to analyze spatial proteomics data with the click of a mouse.
Li says the development of PEELing would only have been possible at Janelia, where biologists can easily collaborate with computer scientists to develop tools that benefit the entire scientific community.
“The primary goal is to help people without a chemical biology background, without a proteomics background, to use our method and to welcome people into this methodology,” Li says. “We need to make this first step simple and robust because people want to focus on their biological discoveries instead of dealing with all the technical details.”
Deciphering signals
Li and his colleagues are also using some of these same methods and tools to learn more about how communication happens inside and outside the cell surface.
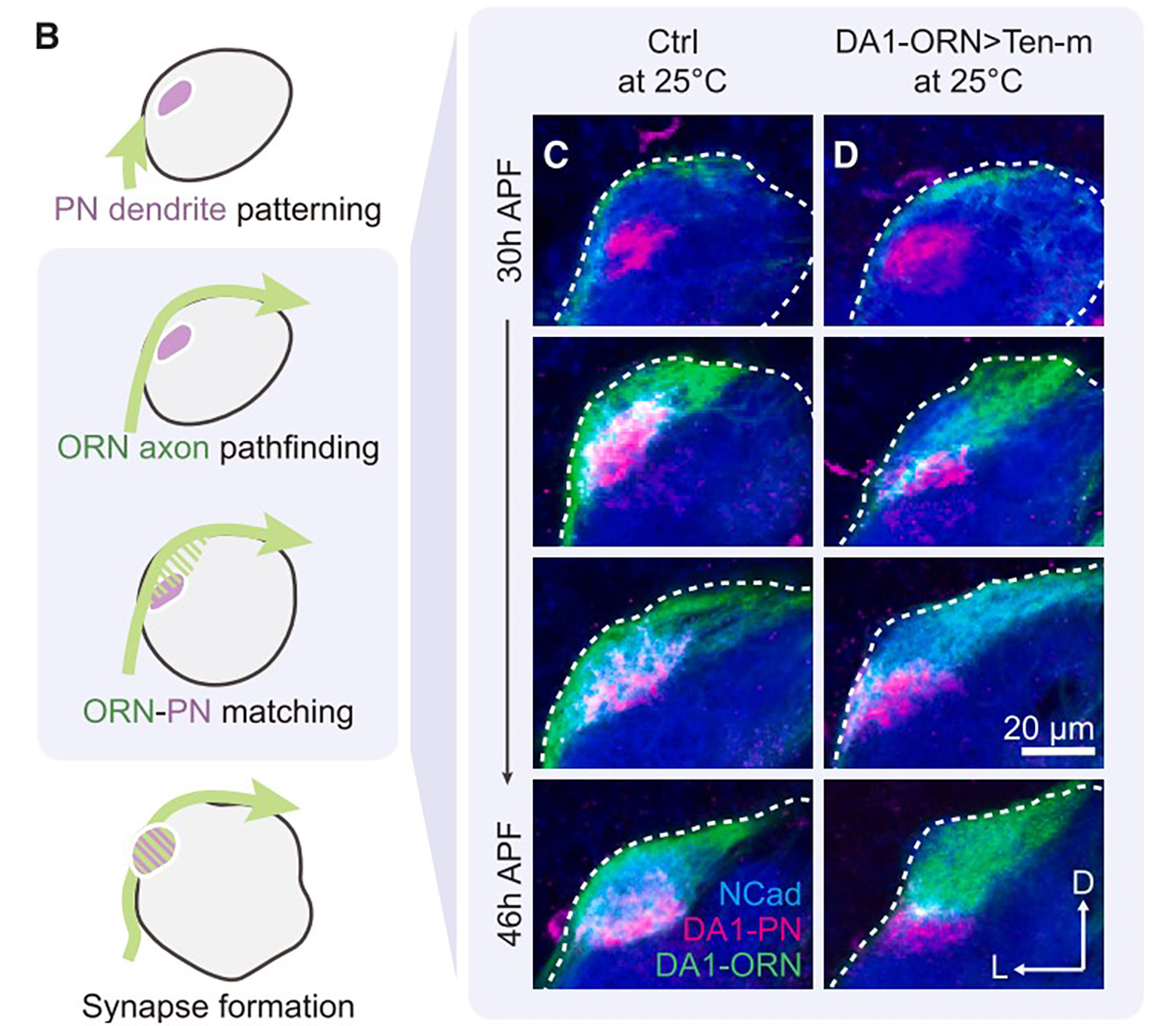
In a recent paper, Li and researchers from Stanford and the Broad Institute examined the signaling that takes place inside neurons after the activation of teneurin, a protein that spans the cell membrane and matches up axons with dendrites to form synapses in the developing brain. Scientists knew that teneurin was responsible for this synaptic matching, but they didn’t know what happened inside the cell after the matching signal was released.
The researchers first used proximity labeling to capture the molecules interacting with teneurin inside the cell. They then used proteomics to identify these proteins and narrow down which ones were potentially involved in the process. The team then used genetics to understand how these candidate proteins interacted with tenurin, enabling the researchers to nail down the signaling partners and elucidate the cellular and molecular mechanisms behind synaptic partner matching.
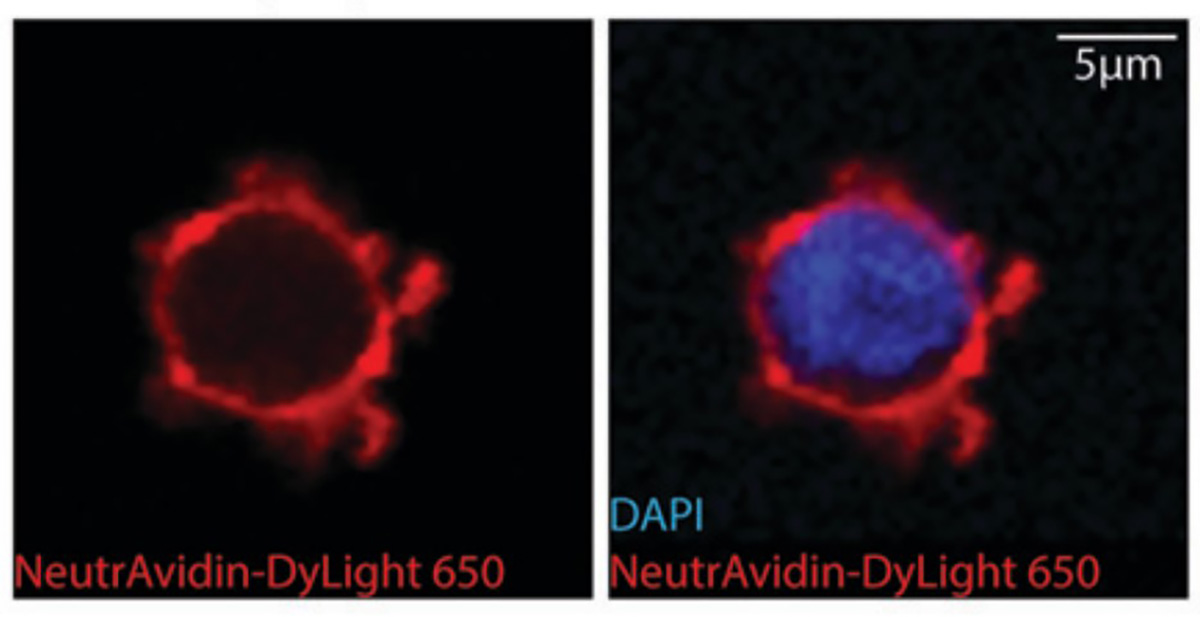
In another recent study, Li and researchers at Stanford and the Broad Institute used cell surface proteomics to examine human dendritic cells, which coordinate the innate and adaptive immune systems via their cell surface proteins. To better understand this coordination, the team looked at how the cell surface changes when dendritic cell activation occurs.
The researchers activated the dendritic cells and then labeled, captured, and quantified the proteins at the cell surface. Next, they annotated these proteins by their known functions and compared the proteins present in activated and resting dendritic cells. Through this characterization, the researchers discovered how the cell modulates the activity of dozens of proteins to regulate the immune response.
“Overall, all of this work comes back to our central goal in the lab to decode the cell surface,” Li says. “Proteomics can provide a comprehensive systems view and really informative hints for follow-up mechanistic studies.”
###
Citations:
Xi Peng, Jody Clements, Zuzhi Jiang, Shuo Han, Stephan Preibisch, Jiefu Li. “PEELing: an integrated and user-centric platform for spatially-resolved proteomics data analysis.” bioRxiv. Posted July 12, 2024. DOI: 10.1101/2023.04.21.537871
Chuanyun Xu, Zhuoran Li, Cheng Lyu, Yixin Hu, Colleen N. McLaughlin, Kenneth Kin Lam Wong, Qijing Xie, David J. Luginbuhl, Hongjie Li, Namrata D. Udeshi, Tanya Svinkina, D.R. Mani, Shuo Han, Tongchao Li, Yang Li, Ricardo Guajardo, Alice Y. Ting, Steven A. Carr, Jiefu Li, Liqun Luo. “Molecular and cellular mechanisms of teneurin signaling in synaptic partner matching.” Cell. Published July 11, 2024. DOI: 10.1016/j.cell.2024.06.022.
Namrata D. Udeshi, Charles Xu, Zuzhi Jiang, Shihong Max Gao, Qian Yin, Wei Luo, Steven A. Carr, Mark M. Davis, Jiefu Li. “Cell-surface Milieu Remodeling in Human Dendritic Cell Activation.” The Journal of Immunology. Published August 12, 2024. DOI: 10.4049/jimmunol.2400089
Media Contacts
Nanci Bompey