Main Menu (Mobile)- Block
- Overview
-
Support Teams
- Overview
- Anatomy and Histology
- Cryo-Electron Microscopy
- Electron Microscopy
- Flow Cytometry
- Gene Targeting and Transgenics
- Immortalized Cell Line Culture
- Integrative Imaging
- Invertebrate Shared Resource
- Janelia Experimental Technology
- Mass Spectrometry
- Media Prep
- Molecular Genomics
- Primary & iPS Cell Culture
- Project Pipeline Support
- Project Technical Resources
- Quantitative Genomics
- Scientific Computing Software
- Scientific Computing Systems
- Viral Tools
- Vivarium
- Open Science
- You + Janelia
- About Us
Main Menu - Block
- Overview
- Anatomy and Histology
- Cryo-Electron Microscopy
- Electron Microscopy
- Flow Cytometry
- Gene Targeting and Transgenics
- Immortalized Cell Line Culture
- Integrative Imaging
- Invertebrate Shared Resource
- Janelia Experimental Technology
- Mass Spectrometry
- Media Prep
- Molecular Genomics
- Primary & iPS Cell Culture
- Project Pipeline Support
- Project Technical Resources
- Quantitative Genomics
- Scientific Computing Software
- Scientific Computing Systems
- Viral Tools
- Vivarium
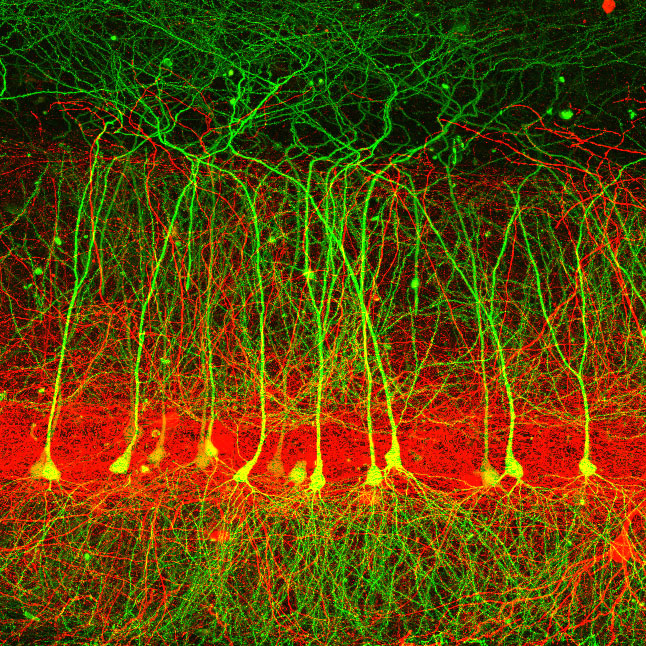
ShuTu Open-Source Software for Neuron Reconstruction
ShuTu (Chinese for “dendrite”) is a software platform for semi-automated reconstruction of neuronal morphology. It is designed for neurons stained following patch-clamp recording and biocytin filling/staining. It can also handle fluorescence images from confocal stacks, however, by first inverting the images.
Even when there is only a single neuron in the imaged volume, automated reconstruction is tricky because of background. Thus, ShuTu was developed to facilitate a two-step process: sophisticated algorithms perform the automated reconstruction, which is then superimposed on the images, along with a convenient user interface to facilitate efficient completion of the reconstruction via manual annotation.
ShuTu was written by Dezhe Jin (Penn State University) and Ting Zhao (Janelia) in collaboration with Nelson Spruston (Janelia).
A more detailed description of ShuTu can be found here.
Tutorial
Installation
To install on Mac and Linux, download the zip file, and in a terminal and in the directory ShuTuMac or ShuTuUbuntu, type the commands
- sudo sh build.sh
- bash
- During the process, enter "yes" or "y" or "Y" when prompted.
On Windows 10, unzip ShuTuWin64.zip to get the GUI software. To install programs for image processing and automatic tracing, install Ubuntu app in the App Store. Here are the steps:
- Update Windows 10 to the current version. Ubuntu app requires Windows 10 version 16215 and up.
- From App Store, install Ubuntu app.
- Use "Turn Windows features on or off," click on "Windows Subsystem for Linux," click OK, reboot.
- Start Ubuntu app.
- sudo apt-get install unzip
- wget http://personal.psu.edu/dzj2/ShuTu/ShuTuUbuntu.zip
- unzip ShuTuUbuntu.zip
- cd ShuTuUbuntu
- sudo sh build.sh
- bash
View source code and collaborate on Github.
Images
Download images for practicing reconstruction.
- Rat dente gyrus granule cell 100X
- Rat dente gyrus granule cell 100X compressed
- Rat dente gyrus granule cell 63X
More Data
Below are some additional images and reconstructions of the neurons described in the paper.