Filter
Associated Lab
- Aguilera Castrejon Lab (2) Apply Aguilera Castrejon Lab filter
- Ahrens Lab (61) Apply Ahrens Lab filter
- Aso Lab (42) Apply Aso Lab filter
- Baker Lab (19) Apply Baker Lab filter
- Betzig Lab (103) Apply Betzig Lab filter
- Beyene Lab (10) Apply Beyene Lab filter
- Bock Lab (14) Apply Bock Lab filter
- Branson Lab (51) Apply Branson Lab filter
- Card Lab (37) Apply Card Lab filter
- Cardona Lab (45) Apply Cardona Lab filter
- Chklovskii Lab (10) Apply Chklovskii Lab filter
- Clapham Lab (14) Apply Clapham Lab filter
- Cui Lab (19) Apply Cui Lab filter
- Darshan Lab (8) Apply Darshan Lab filter
- Dennis Lab (1) Apply Dennis Lab filter
- Dickson Lab (32) Apply Dickson Lab filter
- Druckmann Lab (21) Apply Druckmann Lab filter
- Dudman Lab (42) Apply Dudman Lab filter
- Eddy/Rivas Lab (30) Apply Eddy/Rivas Lab filter
- Egnor Lab (4) Apply Egnor Lab filter
- Espinosa Medina Lab (19) Apply Espinosa Medina Lab filter
- Feliciano Lab (12) Apply Feliciano Lab filter
- Fetter Lab (31) Apply Fetter Lab filter
- FIB-SEM Technology (1) Apply FIB-SEM Technology filter
- Fitzgerald Lab (16) Apply Fitzgerald Lab filter
- Freeman Lab (15) Apply Freeman Lab filter
- Funke Lab (43) Apply Funke Lab filter
- Gonen Lab (59) Apply Gonen Lab filter
- Grigorieff Lab (34) Apply Grigorieff Lab filter
- Harris Lab (55) Apply Harris Lab filter
- Heberlein Lab (13) Apply Heberlein Lab filter
- Hermundstad Lab (26) Apply Hermundstad Lab filter
- Hess Lab (76) Apply Hess Lab filter
- Ilanges Lab (3) Apply Ilanges Lab filter
- Jayaraman Lab (44) Apply Jayaraman Lab filter
- Ji Lab (33) Apply Ji Lab filter
- Johnson Lab (1) Apply Johnson Lab filter
- Karpova Lab (13) Apply Karpova Lab filter
- Keleman Lab (8) Apply Keleman Lab filter
- Keller Lab (61) Apply Keller Lab filter
- Koay Lab (3) Apply Koay Lab filter
- Lavis Lab (144) Apply Lavis Lab filter
- Lee (Albert) Lab (29) Apply Lee (Albert) Lab filter
- Leonardo Lab (19) Apply Leonardo Lab filter
- Li Lab (6) Apply Li Lab filter
- Lippincott-Schwartz Lab (108) Apply Lippincott-Schwartz Lab filter
- Liu (Yin) Lab (3) Apply Liu (Yin) Lab filter
- Liu (Zhe) Lab (60) Apply Liu (Zhe) Lab filter
- Looger Lab (137) Apply Looger Lab filter
- Magee Lab (31) Apply Magee Lab filter
- Menon Lab (12) Apply Menon Lab filter
- Murphy Lab (6) Apply Murphy Lab filter
- O'Shea Lab (6) Apply O'Shea Lab filter
- Otopalik Lab (1) Apply Otopalik Lab filter
- Pachitariu Lab (40) Apply Pachitariu Lab filter
- Pastalkova Lab (5) Apply Pastalkova Lab filter
- Pavlopoulos Lab (7) Apply Pavlopoulos Lab filter
- Pedram Lab (4) Apply Pedram Lab filter
- Podgorski Lab (16) Apply Podgorski Lab filter
- Reiser Lab (49) Apply Reiser Lab filter
- Riddiford Lab (20) Apply Riddiford Lab filter
- Romani Lab (39) Apply Romani Lab filter
- Rubin Lab (111) Apply Rubin Lab filter
- Saalfeld Lab (47) Apply Saalfeld Lab filter
- Satou Lab (3) Apply Satou Lab filter
- Scheffer Lab (38) Apply Scheffer Lab filter
- Schreiter Lab (53) Apply Schreiter Lab filter
- Sgro Lab (2) Apply Sgro Lab filter
- Shroff Lab (31) Apply Shroff Lab filter
- Simpson Lab (18) Apply Simpson Lab filter
- Singer Lab (37) Apply Singer Lab filter
- Spruston Lab (61) Apply Spruston Lab filter
- Stern Lab (75) Apply Stern Lab filter
- Sternson Lab (47) Apply Sternson Lab filter
- Stringer Lab (38) Apply Stringer Lab filter
- Svoboda Lab (132) Apply Svoboda Lab filter
- Tebo Lab (11) Apply Tebo Lab filter
- Tervo Lab (9) Apply Tervo Lab filter
- Tillberg Lab (19) Apply Tillberg Lab filter
- Tjian Lab (17) Apply Tjian Lab filter
- Truman Lab (58) Apply Truman Lab filter
- Turaga Lab (41) Apply Turaga Lab filter
- Turner Lab (27) Apply Turner Lab filter
- Vale Lab (8) Apply Vale Lab filter
- Voigts Lab (4) Apply Voigts Lab filter
- Wang (Meng) Lab (27) Apply Wang (Meng) Lab filter
- Wang (Shaohe) Lab (6) Apply Wang (Shaohe) Lab filter
- Wong-Campos Lab (1) Apply Wong-Campos Lab filter
- Wu Lab (8) Apply Wu Lab filter
- Zlatic Lab (26) Apply Zlatic Lab filter
- Zuker Lab (5) Apply Zuker Lab filter
Associated Project Team
- CellMap (12) Apply CellMap filter
- COSEM (3) Apply COSEM filter
- FIB-SEM Technology (5) Apply FIB-SEM Technology filter
- Fly Descending Interneuron (12) Apply Fly Descending Interneuron filter
- Fly Functional Connectome (14) Apply Fly Functional Connectome filter
- Fly Olympiad (5) Apply Fly Olympiad filter
- FlyEM (56) Apply FlyEM filter
- FlyLight (50) Apply FlyLight filter
- GENIE (47) Apply GENIE filter
- Integrative Imaging (9) Apply Integrative Imaging filter
- Larval Olympiad (2) Apply Larval Olympiad filter
- MouseLight (18) Apply MouseLight filter
- NeuroSeq (1) Apply NeuroSeq filter
- ThalamoSeq (1) Apply ThalamoSeq filter
- Tool Translation Team (T3) (29) Apply Tool Translation Team (T3) filter
- Transcription Imaging (45) Apply Transcription Imaging filter
Associated Support Team
- Project Pipeline Support (5) Apply Project Pipeline Support filter
- Anatomy and Histology (18) Apply Anatomy and Histology filter
- Cryo-Electron Microscopy (41) Apply Cryo-Electron Microscopy filter
- Electron Microscopy (18) Apply Electron Microscopy filter
- Gene Targeting and Transgenics (11) Apply Gene Targeting and Transgenics filter
- High Performance Computing (7) Apply High Performance Computing filter
- Integrative Imaging (19) Apply Integrative Imaging filter
- Invertebrate Shared Resource (40) Apply Invertebrate Shared Resource filter
- Janelia Experimental Technology (37) Apply Janelia Experimental Technology filter
- Management Team (1) Apply Management Team filter
- Mass Spectrometry (1) Apply Mass Spectrometry filter
- Molecular Genomics (15) Apply Molecular Genomics filter
- Project Technical Resources (54) Apply Project Technical Resources filter
- Quantitative Genomics (20) Apply Quantitative Genomics filter
- Scientific Computing (103) Apply Scientific Computing filter
- Stem Cell & Primary Culture (14) Apply Stem Cell & Primary Culture filter
- Viral Tools (14) Apply Viral Tools filter
- Vivarium (7) Apply Vivarium filter
Publication Date
- 2026 (31) Apply 2026 filter
- 2025 (223) Apply 2025 filter
- 2024 (211) Apply 2024 filter
- 2023 (157) Apply 2023 filter
- 2022 (166) Apply 2022 filter
- 2021 (175) Apply 2021 filter
- 2020 (177) Apply 2020 filter
- 2019 (177) Apply 2019 filter
- 2018 (206) Apply 2018 filter
- 2017 (186) Apply 2017 filter
- 2016 (191) Apply 2016 filter
- 2015 (195) Apply 2015 filter
- 2014 (190) Apply 2014 filter
- 2013 (136) Apply 2013 filter
- 2012 (112) Apply 2012 filter
- 2011 (98) Apply 2011 filter
- 2010 (61) Apply 2010 filter
- 2009 (56) Apply 2009 filter
- 2008 (40) Apply 2008 filter
- 2007 (21) Apply 2007 filter
- 2006 (3) Apply 2006 filter
2812 Janelia Publications
Showing 1171-1180 of 2812 resultsThe identification of synaptic partners is challenging in dense nerve bundles, where many processes occupy regions beneath the resolution of conventional light microscopy. To address this difficulty, we have developed GRASP, a system to label membrane contacts and synapses between two cells in living animals. Two complementary fragments of GFP are expressed on different cells, tethered to extracellular domains of transmembrane carrier proteins. When the complementary GFP fragments are fused to ubiquitous transmembrane proteins, GFP fluorescence appears uniformly along membrane contacts between the two cells. When one or both GFP fragments are fused to synaptic transmembrane proteins, GFP fluorescence is tightly localized to synapses. GRASP marks known synaptic contacts in C. elegans, correctly identifies changes in mutants with altered synaptic specificity, and can uncover new information about synaptic locations as confirmed by electron microscopy. GRASP may prove particularly useful for defining connectivity in complex nervous systems.
Proper functioning of organelles such as the ER or the Golgi apparatus requires luminal accumulation of Ca(2+) at high concentrations. Here we describe a ratiometric low-affinity Ca(2+) sensor of the GFP-aequorin protein (GAP) family optimized for measurements in high-Ca(2+) concentration environments. Transgenic animals expressing the ER-targeted sensor allowed monitoring of Ca(2+) signals inside the organelle. The use of the sensor was demonstrated under three experimental paradigms: (1) ER Ca(2+) oscillations in cultured astrocytes, (2) ex vivo functional mapping of cholinergic receptors triggering ER Ca(2+) release in acute hippocampal slices from transgenic mice, and (3) in vivo sarcoplasmic reticulum Ca(2+) dynamics in the muscle of transgenic flies. Our results provide proof of the suitability of the new biosensors to monitor Ca(2+) dynamics inside intracellular organelles under physiological conditions and open an avenue to explore complex Ca(2+) signaling in animal models of health and disease.
When a behavior repeatedly fails to achieve its goal, animals often give up and become passive, which can be strategic for preserving energy or regrouping between attempts. It is unknown how the brain identifies behavioral failures and mediates this behavioral-state switch. In larval zebrafish swimming in virtual reality, visual feedback can be withheld so that swim attempts fail to trigger expected visual flow. After tens of seconds of such motor futility, animals became passive for similar durations. Whole-brain calcium imaging revealed noradrenergic neurons that responded specifically to failed swim attempts and radial astrocytes whose calcium levels accumulated with increasing numbers of failed attempts. Using cell ablation and optogenetic or chemogenetic activation, we found that noradrenergic neurons progressively activated brainstem radial astrocytes, which then suppressed swimming. Thus, radial astrocytes perform a computation critical for behavior: they accumulate evidence that current actions are ineffective and consequently drive changes in behavioral states.
We found that glia secrete myoglianin, a TGF-β ligand, to instruct developmental neural remodeling in Drosophila. Glial myoglianin upregulated neuronal expression of an ecdysone nuclear receptor that triggered neurite remodeling following the late-larval ecdysone peak. Thus glia orchestrate developmental neural remodeling not only by engulfment of unwanted neurites but also by enabling neuron remodeling.
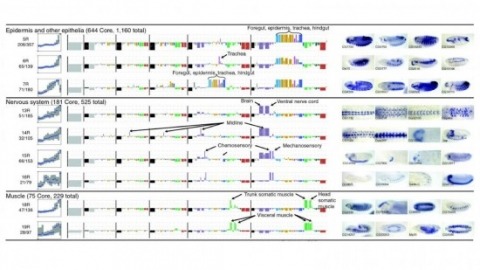
Cell and tissue specific gene expression is a defining feature of embryonic development in multi-cellular organisms. However, the range of gene expression patterns, the extent of the correlation of expression with function, and the classes of genes whose spatial expression are tightly regulated have been unclear due to the lack of an unbiased, genome-wide survey of gene expression patterns.
Neuroscience research is becoming increasingly more collaborative and interdisciplinary with partnerships between industry and academia and insights from fields beyond neuroscience. In the age of institutional initiatives and multi-investigator collaborations, scientists from around the world shared their perspectives on the effectiveness of large-scale collaborations versus single-lab, hypothesis-driven science.
Connectomics is a subfield of neuroscience that aims to map the brain’s intricate wiring diagram. Accurate neuron segmentation from microscopy volumes is essential for automating connectome reconstruction. However, current state-of-the-art algorithms use image-based convolutional neural networks that are limited to local neuron shape context. Thus, we introduce a new framework that reasons over global neuron shape with a novel point affinity transformer. Our framework embeds a (multi-)neuron point cloud into a fixed-length feature set from which we can decode any point pair affinities, enabling clustering neuron point clouds for automatic proofreading. We also show that the learned feature set can easily be mapped to a contrastive embedding space that enables neuron type classification using a simple KNN classifier. Our approach excels in two demanding connectomics tasks: proofreading segmentation errors and classifying neuron types. Evaluated on three benchmark datasets derived from state-of-the-art connectomes, our method outperforms point transformers, graph neural networks, and unsupervised clustering baselines.
The fluorescent glutamate indicator iGluSnFR enables imaging of neurotransmission with genetic and molecular specificity. However, existing iGluSnFR variants exhibit low in vivo signal-to-noise ratios, saturating activation kinetics and exclusion from postsynaptic densities. Using a multiassay screen in bacteria, soluble protein and cultured neurons, we generated variants with improved signal-to-noise ratios and kinetics. We developed surface display constructs that improve iGluSnFR's nanoscopic localization to postsynapses. The resulting indicator iGluSnFR3 exhibits rapid nonsaturating activation kinetics and reports synaptic glutamate release with decreased saturation and increased specificity versus extrasynaptic signals in cultured neurons. Simultaneous imaging and electrophysiology at individual boutons in mouse visual cortex showed that iGluSnFR3 transients report single action potentials with high specificity. In vibrissal sensory cortex layer 4, we used iGluSnFR3 to characterize distinct patterns of touch-evoked feedforward input from thalamocortical boutons and both feedforward and recurrent input onto L4 cortical neuron dendritic spines.
Identifying the input-output operations of neurons requires measurements of synaptic transmission simultaneously at many of a neuron’s thousands of inputs in the intact brain. To facilitate this goal, we engineered and screened 3365 variants of the fluorescent protein glutamate indicator iGluSnFR3 in neuron culture, and selected variants in the mouse visual cortex. Two variants have high sensitivity, fast activation (< 2 ms) and deactivation times tailored for recording large populations of synapses (iGluSnFR4s, 153 ms) or rapid dynamics (iGluSnFR4f, 26 ms). By imaging action-potential evoked signals on axons and visually-evoked signals on dendritic spines, we show that iGluSnFR4s/4f primarily detect local synaptic glutamate with single-vesicle sensitivity. The indicators detect a wide range of naturalistic synaptic transmission, including in the vibrissal cortex layer 4 and in hippocampal CA1 dendrites. iGluSnFR4 increases the sensitivity and scale (4s) or speed (4f) of tracking information flow in neural networks in vivo.
Understanding how neurons integrate signals from thousands of input synapses requires methods to monitor neurotransmission across many sites simultaneously. The fluorescent protein glutamate indicator iGluSnFR enables visualization of synaptic signaling, but the sensitivity, scale and speed of such measurements are limited by existing variants. Here we developed two highly sensitive fourth-generation iGluSnFR variants with fast activation and tailored deactivation rates: iGluSnFR4f for tracking rapid dynamics, and iGluSnFR4s for recording from large populations of synapses. These indicators detect glutamate with high spatial specificity and single-vesicle sensitivity in vivo. We used them to record natural patterns of synaptic transmission across multiple experimental contexts in mice, including two-photon imaging in cortical layers 1–4 and hippocampal CA1, and photometry in the midbrain. The iGluSnFR4 variants extend the speed, sensitivity and scalability of glutamate imaging, enabling direct observation of information flow through neural networks in the intact brain.