To an outside observer, many scientific images appear to be snapshots – still-frames of fluorescently labeled neurons, for instance, or the arcane structure of a molecule. But in reality, these images are a digest of colossal datasets. Hundreds of thousands of images (in extreme cases, upwards of a million) are compiled, layer by layer, to generate a single image that deepens scientific insight.
How are such copious amounts of information processed and stored? At Janelia, the Scientific Computing department devises, implements, and maintains the strategies that help scientists meet this challenge.
The power of the Scientific Computing support team is twofold – hardware and software . Top-of-the-line infrastructure provides robust computing power and nearly five petabytes of storage (1.5 petabytes is roughly equivalent to 10 billion photos on Facebook), while engineers collaborate with Janelia scientists to design software-based solutions to scientific obstacles.
While the support team’s software engineers mainly collaborate with project teams and a handful of individual labs, nearly everyone takes advantage of the team’s computing gem, “the Cluster.”
Group Leader and structural biologist Nikolaus Grigorieff, for example, says that without the Cluster, processing the data reported in a recent publication would have taken more than a year longer. Grigorieff’s paper detailed viral mRNA translation by capturing successive phases of ribosomal interaction. “It was one of the bigger datasets I’ve collected, over a million ribosome images,” he says.
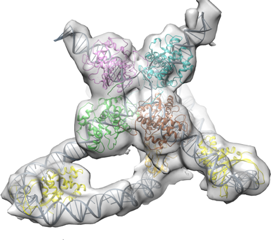
Using the computing power from the Cluster, Nikolas Grigorieff can efficiently process data needed to create structural images, like the above Holliday Junction, a protein complex that aids in genetic recombination, a process that generates genetic diversity. Image courtesy of Niko Grigorieff
Grigorieff explains that the Cluster’s data-crunching power is not so mysterious – there’s strength in numbers. Because the computing system contains some 5,000 cores, which are akin to central processing units (CPUs), 5,000 independent pieces of work can be run simultaneously, making data processing thousands of times faster than a single computer.
But for scientists like Group Leader Tamir Gonen, computing power is not the battle at hand. Gonen devised a new cryoEM technique, called Micro electron diffraction, or MicroED, that addresses a problematic bottleneck in traditional protein-structure imaging. The old standby, X-ray crystallography, requires relatively large crystals for successful imaging.
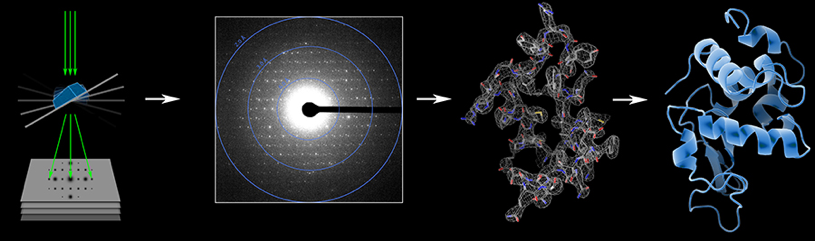
Tamir Gonen developed the MicroED technique, illustrated here, to image tiny crystals using electron diffraction. The resulting data helps him decipher the structure of proteins. Image courtesy of Tamir Gonen
“Every large crystal was once a small crystal, but not every small crystal can become a large crystal,” Gonen explains. With Gonen’s MicroED technique, he and his colleagues have been able to image small, bountiful crystals that are unfit for X-ray diffraction (they are 1 billion times smaller in volume than required). Imaging data in hand, Gonen began to brainstorm about processing strategies.
“Instead of developing new software to process raw MicroED data, we wanted to leverage the software developed for X-ray crystallography over the past 40 years,” says Gonen. After Gonen consulted with the Scientific Computing team, the team’s engineers generated a pipeline that tricked the X-ray diffraction software into reading MicroED datasets. Without the translational software, Gonen says, the project might still be two decades away from true scientific impact.
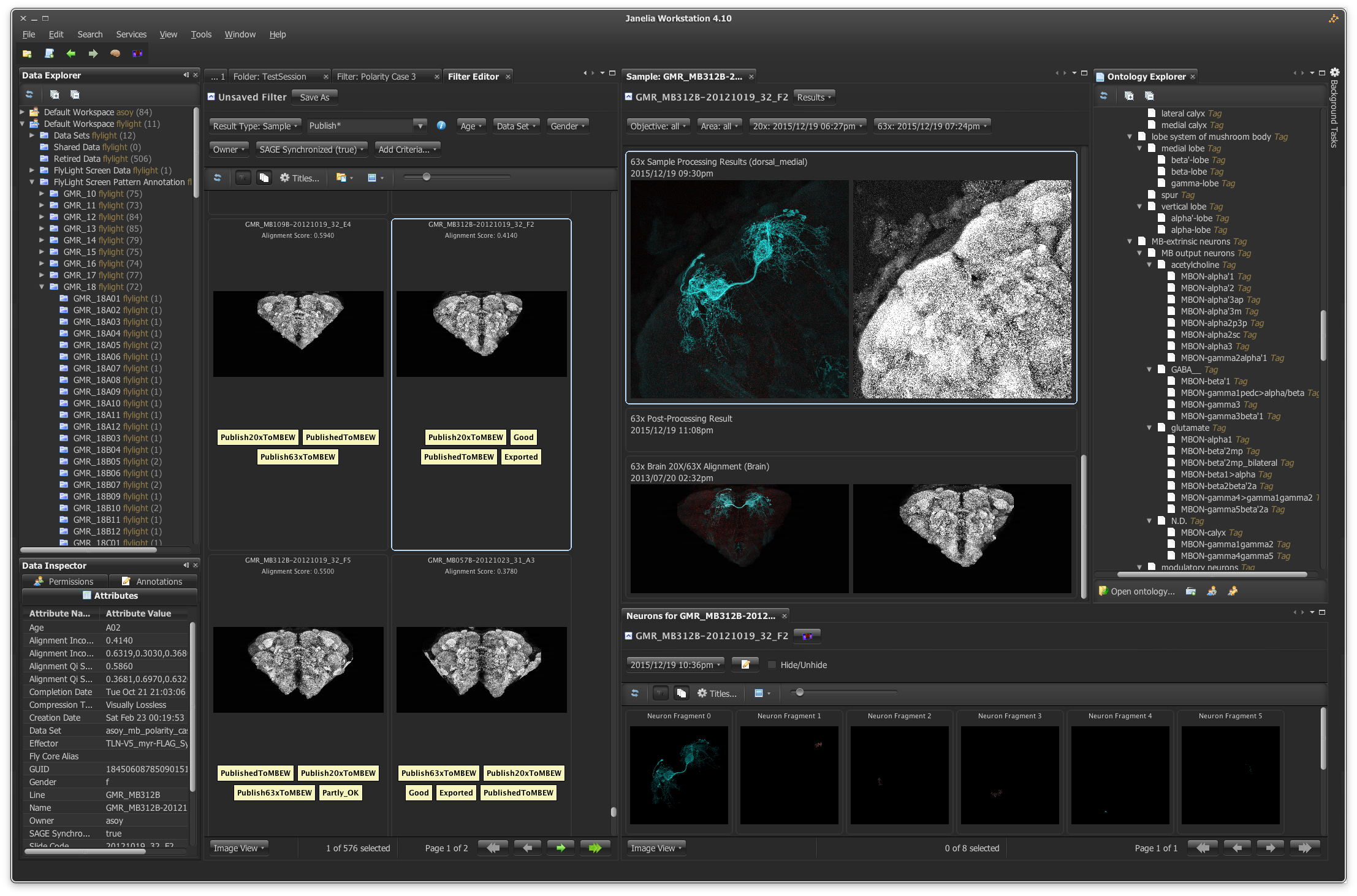
In another example of project catalysis, Sean Murphy, manager of scientific software at Janelia, joined the MouseLight project team early on and helped define its goals. Led by Jayaram Chandrashekar, the MouseLight team annotates and traces complete neuronal pathways in the mouse brain, with the ultimate goal of delineating a carefully selected group of about 1,000 neurons. To reconstruct neurons in 3D, the scientists must capture hundreds of thousands of high-resolution images, generating huge datasets.
Using a combination of custom in-house designed software programs, created by Sean Murphy, the MouseLight team traces neurons at multiple Janelia workstations. Image courtesy of Sean Murphy
“A person can look at and assess a lot of data, but in this case, the scientist has to go through tens of terabytes of data to trace these neurons,” says Murphy. “How do we pump that much imagery through their visual system in a timely manner?”
Murphy and a team of developers answered that challenge with specially designed software that compiles the data from each brain sample into information-rich 3D images, allowing scientists to trace and annotate the trajectory of a neuron in a matter of days.
“Before, the team would have had to go through the data stocks manually, with poorly matched software, maybe tracing one neuron every 6 months,” says Murphy. Now, tracing a neuron takes a person several days. Murphy and the Scientific Computing team are currently working with the MouseLight project team to automate the process and, ideally, cut that time down by an order of magnitude.
Stories of Collaboration
Collaboration between labs, project teams, support teams, and scientists at other institutions is an essential part of the culture and intellectual life at Janelia.


