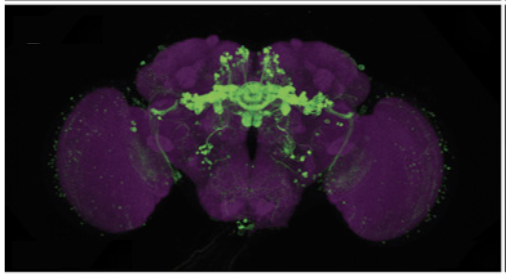
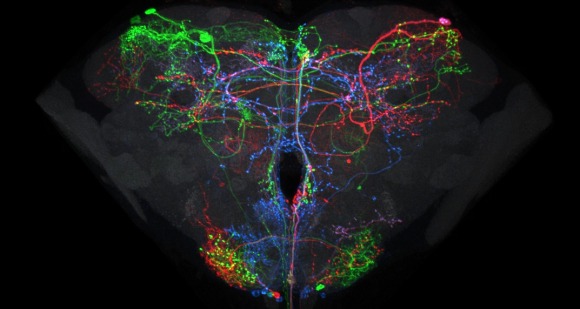
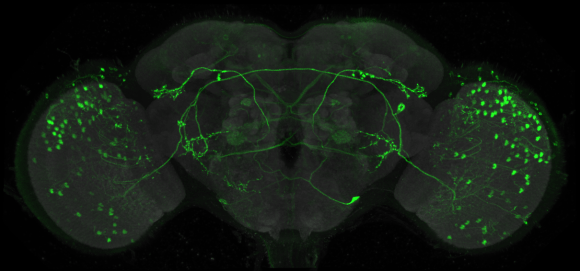
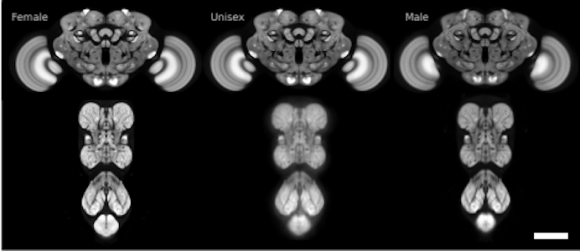
The FlyLight Project produces large anatomical data sets and highly characterized collections of GAL4, LexA and Split-GAL4 drivers in order to enable the visualization and precise manipulation of individual cell types in the Drosophila nervous system.
News & Updates
FlyLight’s goal is to generate, with the help of collaborators, a set of split-GAL4 lines that specifically express in the vast majority of cell types of the fly nervous system.
FlyLight makes its primary data available through websites for expression patterns: Generation 1 GAL4 and LexA lines, Generation 1 MultiColor FlpOut, and Split-GAL4 lines. NeuronBridge allows anatomical searching between Janelia's light and electron microscopy datasets.
Bloomington Drosophila Stock Center distributes Generation 1, split-hemidriver, and many split-GAL4 combination lines. Vienna Drosophila Resource Center distributes VT Generation 1 lines.
To request other split-GAL4 lines or for questions, contact us at flylight@janelia.hhmi.org.
Left. Diagram of a portion of a neural circuit with one cell labeled by expression of a GAL4 driver (green). Right. Comparison of light and electron microscopic (EM) morphologies establishes the correspondence between nodes in the connectome and the GAL4 drivers that express in them.
At Janelia, we believe that the unparalleled experimental versatility of Drosophila provides an opportunity for an intellectual approach to understanding neuronal circuits not now feasible in vertebrate systems. The modest size of the fly’s nervous system—100,000 neurons in the adult and 10,000 in the larva—allows a systematic approach to understanding the complete nervous system of an organism that exhibits a fascinating repertoire of behaviors. A detailed wiring diagram is essential, but not sufficient, to understand the function of a neural network. We need to be able to point to any node in that wiring diagram and be able to silence or activate that node or more subtly modify its properties—for example, by inactivating a particular neuromodulator receptor in just that node. We need to be able to measure the effects of those manipulations on the behavior of the animal, the ensemble activity of the local neural network and the activity of individual neurons—a neuron or a network that can be reliably identified in a different animal, tomorrow or a year from now.
FlyLight plays a key role in this strategy by providing the bridge from electron-microscopic-level anatomical studies to behavioral and physiological functional studies. Cell-type-specific GAL4 drivers can be used to characterize the morphology of individual cells using the stochastic labeling methods we developed for this project. Matching the shape of a neuron determined in this way to a cell reconstructed from serial electron micrographs can reliably establish the correspondence between a node in the connectome and driver lines that can be used to express indicators and modifiers of function specifically in that node.
Relating the data from the larval neural atlas (B) to the EM reconstruction of behavioral circuits (A) and the generation of intersection lines targeted to specific neurons (C). A: EM reconstruction of the local and the brain circuit loops that mediate larval rolling behavior in response to mechanosensory stimulation (by C. Schneider-Mizell and A. Cardona). Reconstructed neurons in the short and long loops are in various colors. Inset shows the location of the neurons relative to the outline of the CNS. B: Examples of light-level images of the anatomy of some of the neurons in the circuit based on flip-out analysis of the Janelia GAL4 collection. The color around the name matches the cells in the reconstruction. C: Examples of intersection lines that cleanly target specific neurons in the loop and which evoke rolling behavior when stimulated (studies by T. Oyama and M. Zlatic)
At the heart of the FlyLight project are large-scale data production processes that enable us to highlight and study cells at various points in the life of the fruit fly. We have developed and implemented a number of novel methods for brain dissection, optical clearing of tissue, labeling and automated microscopy. We have also developed computational tools for stitching together multi-stack images, aligning brain images into a common framework, annotating expression patterns, separating individual neurons from multicolor images containing several neurons, and for displaying and manipulating image stacks and other anatomical data.
To establish this functional bridge between the EM-level anatomy and cell-type-specific manipulation and analysis, we:
- Image and annotate several thousand GAL4 and LexA driver lines that label different sets of neurons (Generation 1 lines) in the adult Drosophila and the third instar larva
- Collaborate with world-class anatomical experts to generate a comprehensive library of split-GAL4 driver lines to reproducibly target neurons with cell-type specific resolution.
- Image those lines at high resolution to assemble a complete catalog of single neuron morphologies of the larval and adult Drosophila central nervous system by stochastic labeling of neurons and by imaging reporters targeted to synapses.
- Produce stable fly stocks for further analysis in well-controlled behavioral assays
To find out more, please visit our Research page.