Main Menu (Mobile)- Block
- Overview
-
Support Teams
- Overview
- Anatomy and Histology
- Cryo-Electron Microscopy
- Electron Microscopy
- Flow Cytometry
- Gene Targeting and Transgenics
- High Performance Computing
- Immortalized Cell Line Culture
- Integrative Imaging
- Invertebrate Shared Resource
- Janelia Experimental Technology
- Mass Spectrometry
- Media Prep
- Molecular Genomics
- Stem Cell & Primary Culture
- Project Pipeline Support
- Project Technical Resources
- Quantitative Genomics
- Scientific Computing
- Viral Tools
- Vivarium
- Open Science
- You + Janelia
- About Us
Main Menu - Block
- Overview
- Anatomy and Histology
- Cryo-Electron Microscopy
- Electron Microscopy
- Flow Cytometry
- Gene Targeting and Transgenics
- High Performance Computing
- Immortalized Cell Line Culture
- Integrative Imaging
- Invertebrate Shared Resource
- Janelia Experimental Technology
- Mass Spectrometry
- Media Prep
- Molecular Genomics
- Stem Cell & Primary Culture
- Project Pipeline Support
- Project Technical Resources
- Quantitative Genomics
- Scientific Computing
- Viral Tools
- Vivarium
The Janelia Archives
Artifact Name: IsoView Light-Sheet Microscope Science
Science
The IsoView light-sheet microscope, developed in the lab of Janelia Group Leader Philipp Keller in 2015, produces images of entire organisms, such as a zebrafish or fruit fly embryo, with enough resolution in all three dimensions that each cell appears as a distinct structure. What's more, it does so within a few hundred milliseconds – fast enough to watch cells migrate as a developing embryo takes shape and to monitor brain activity as neuronal circuits fire.
Previous methods enabled researchers to perform fast three-dimensional imaging of large specimens, but internal cellular structures remained inaccessible. With the IsoView, subcellular structures can be distinguished – without sacrificing speed or field of view. Rather than collecting a single image of a sample with a single objective, the new microscope simultaneously captures images from multiple angles. Each image still suffers from poor resolution along one axis, but combining the most useful data from each image generates a final image with 400 nm resolution in all dimensions – 10-fold better than conventional light-sheet microscopy for large-volume imaging.
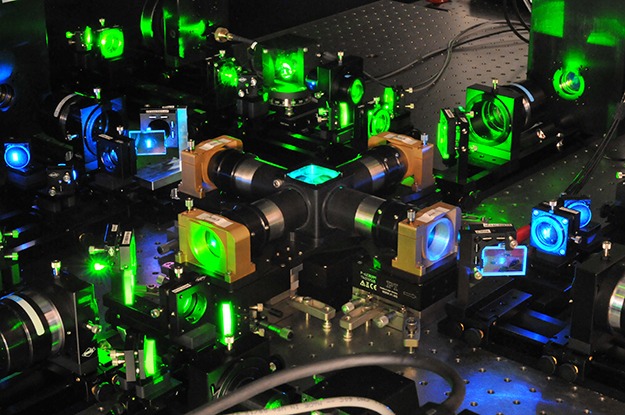
The IsoView framework is the result of collaboration between Janelians Raghav Chhetri, an optical physicist who helped develop the image collection methods, and Fernando Amat, a computer scientist who developed algorithms to process the images. The microscope produces approximately 10 terabytes of multiview image data from one hour of imaging, which required developing an entirely new software pipeline to register and deconvolve the images. The 2015 Nature Methods article describing the microscope includes complete building plans for the microscope and associated image-processing software developed by Keller’s team.
On the left, lateral and ventral views of a Drosophila embryo close to hatching, labelled with the GCaMP family of variously colored, ultrasensitive calcium sensors. The colors represent the developing nervous system and indicate the intensity of calcium signaling (A) and the structure’s depth through the body (B).
On the right, lateral and ventral views of a developing Drosophila embryo. The green color marks cell nuclei and the magenta marks cell membranes. The magenta stripe represents the ventral furrow, which forms during gastrulation.
A time series of the ventral view of a developing Drosophila embryo shows the progression of the ventral furrow during gastrulation. The time series begins approximately 3 hours after egg-laying, and the entire process lasts somewhere between 20-30 minutes. The green color marks cell nuclei and the magenta marks cell membranes.
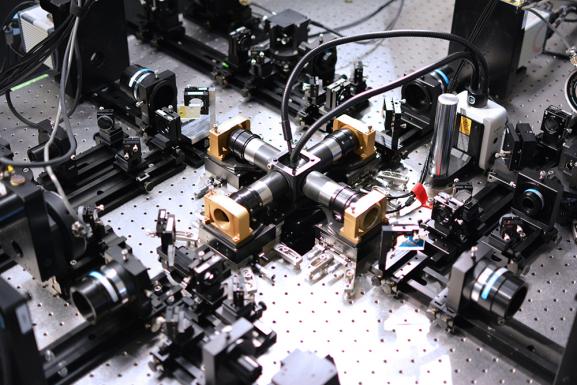
The IsoView lattice light sheet microscope, an isotropic, multi-view microscope, surrounds the specimen to be imaged with four objectives at right angles to one another, each connected to a camera. Green and blue laser beams illuminate the sample for 2-color imaging. The multiple views allow researchers to computationally enhance the image resolution.