Main Menu (Mobile)- Block
- Overview
-
Support Teams
- Overview
- Anatomy and Histology
- Cryo-Electron Microscopy
- Electron Microscopy
- Flow Cytometry
- Gene Targeting and Transgenics
- High Performance Computing
- Immortalized Cell Line Culture
- Integrative Imaging
- Invertebrate Shared Resource
- Janelia Experimental Technology
- Mass Spectrometry
- Media Prep
- Molecular Genomics
- Stem Cell & Primary Culture
- Project Pipeline Support
- Project Technical Resources
- Quantitative Genomics
- Scientific Computing
- Viral Tools
- Vivarium
- Open Science
- You + Janelia
- About Us
Main Menu - Block
- Overview
- Anatomy and Histology
- Cryo-Electron Microscopy
- Electron Microscopy
- Flow Cytometry
- Gene Targeting and Transgenics
- High Performance Computing
- Immortalized Cell Line Culture
- Integrative Imaging
- Invertebrate Shared Resource
- Janelia Experimental Technology
- Mass Spectrometry
- Media Prep
- Molecular Genomics
- Stem Cell & Primary Culture
- Project Pipeline Support
- Project Technical Resources
- Quantitative Genomics
- Scientific Computing
- Viral Tools
- Vivarium
The Janelia Archives
Artifact Name: Micro-Electron Diffraction (MicroED) Science
Science
Before scientists can design drugs to target a disease-causing protein, they have to resolve the protein’s structure. To date, researchers have typically determined protein structures by x-ray crystallography, which requires precipitating the proteins into large crystals, exposing the crystals to x-rays, and analyzing the x-ray diffraction patterns. The crystals must be large enough to withstand the damage caused by x-rays. Many proteins, however, don’t crystallize easily, or at all.
To circumvent this problem, Tamir Gonen’s research group at Janelia developed a method called micro-electron diffraction, or MicroED, which enabled them to determine the structures of proteins from submicron-sized crystals. The method involves shooting electrons at tiny protein crystals under cryogenic conditions and collecting the resulting electron diffraction patterns. Electrons interact with matter more strongly than x-rays, which is why it’s possible to obtain useful data from very tiny crystals.
A single electron diffraction pattern is insufficient to determine a protein’s structure, but by gradually rotating a tiny crystal repeatedly before shooting it again with electrons, and thus collecting diffraction patterns from multiple angles, it becomes possible to mathematically reconstruct the protein structure. Gonen and his team solved the structure of lysozyme using this method, and published their findings in eLife in 2013. Since then, Gonen’s team has determined many other structures, and the method has been adopted by a number of other laboratories around the world.
A cartoon representation of the MicroED technique, in which electron beams (green) are aimed at a tiny protein crystal, with the resulting diffraction pattern collected for later analysis. The crystal is rotated one degree at a time, and diffraction patterns are collected at each new angle.
An example of a MicroED-generated electron diffraction pattern.
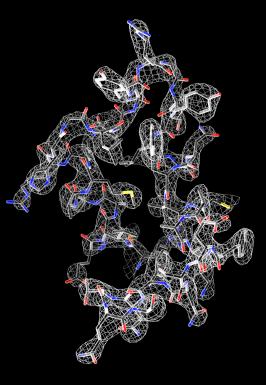
Raw data representation of the structure of lysozyme, the first protein structure ever determined by MicroED. The structure was published in eLife in 2013.
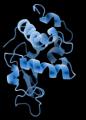
The final protein structure model of lysozyme, as determined by MicroED.
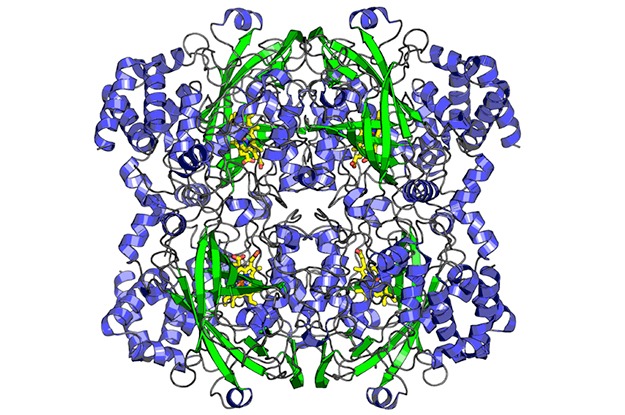
The final structure of catalase, the second structure ever determined by MicroED. The structure was solved from a single nanocrystal that was less than 200 nm thick. The structure was published in eLife in 2014.