Select Publications
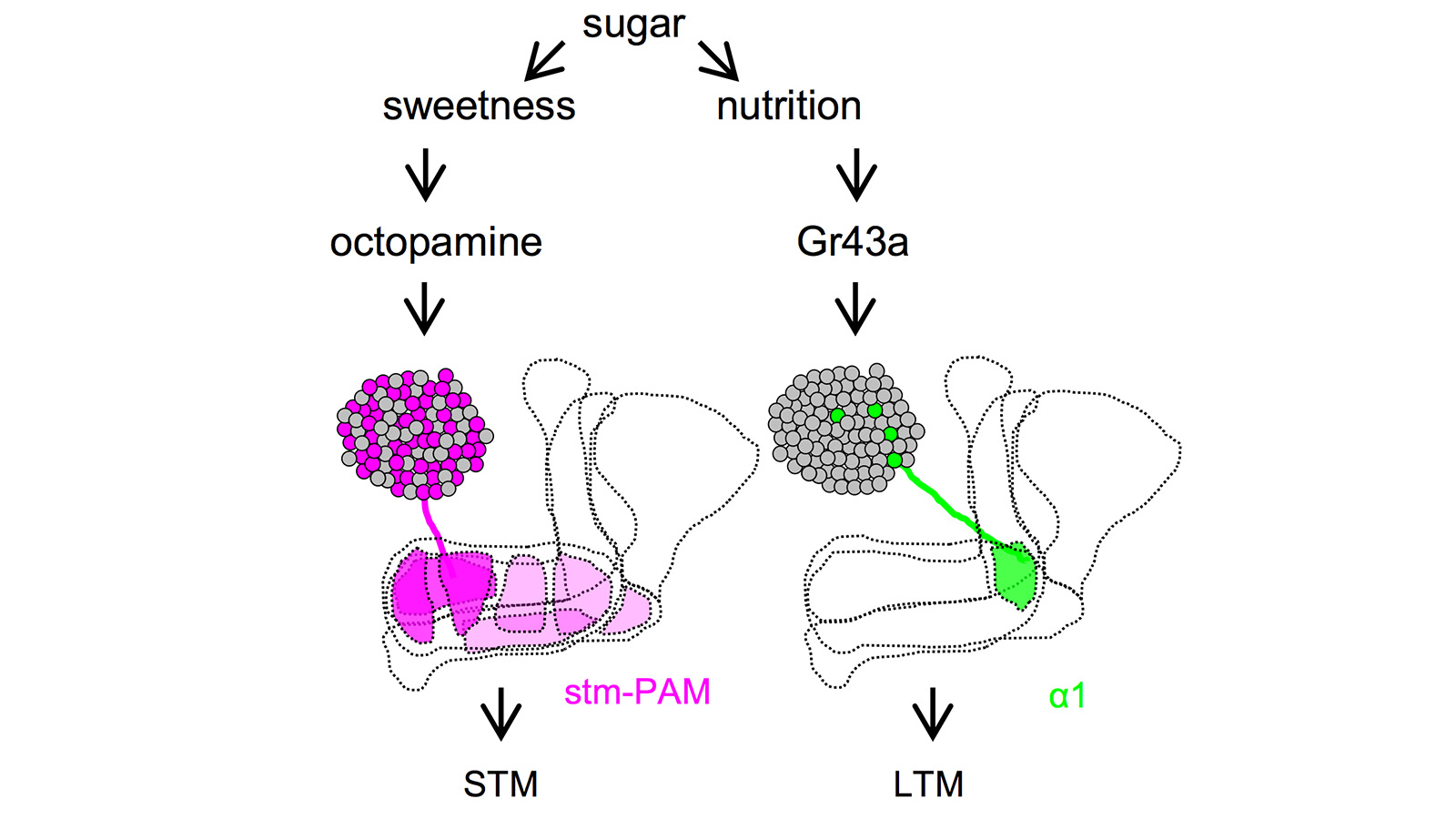
View More PublicationsDrosophila melanogaster can acquire a stable appetitive olfactory memory when the presentation of a sugar reward and an odor are paired. However, the neuronal mechanisms by which a single training induces long-term memory are poorly understood. Here we show that two distinct subsets of dopamine neurons in the fly brain signal reward for short-term (STM) and long-term memories (LTM). One subset induces memory that decays within several hours, whereas the other induces memory that gradually develops after training. They convey reward signals to spatially segregated synaptic domains of the mushroom body (MB), a potential site for convergence. Furthermore, we identified a single type of dopamine neuron that conveys the reward signal to restricted subdomains of the mushroom body lobes and induces long-term memory. Constant appetitive memory retention after a single training session thus comprises two memory components triggered by distinct dopamine neurons.
View Publication PageAnimals discriminate stimuli, learn their predictive value and use this knowledge to modify their behavior. In Drosophila, the mushroom body (MB) plays a key role in these processes. Sensory stimuli are sparsely represented by ∼2000 Kenyon cells, which converge onto 34 output neurons (MBONs) of 21 types. We studied the role of MBONs in several associative learning tasks and in sleep regulation, revealing the extent to which information flow is segregated into distinct channels and suggesting possible roles for the multi-layered MBON network. We also show that optogenetic activation of MBONs can, depending on cell type, induce repulsion or attraction in flies. The behavioral effects of MBON perturbation are combinatorial, suggesting that the MBON ensemble collectively represents valence. We propose that local, stimulus-specific dopaminergic modulation selectively alters the balance within the MBON network for those stimuli. Our results suggest that valence encoded by the MBON ensemble biases memory-based action selection.
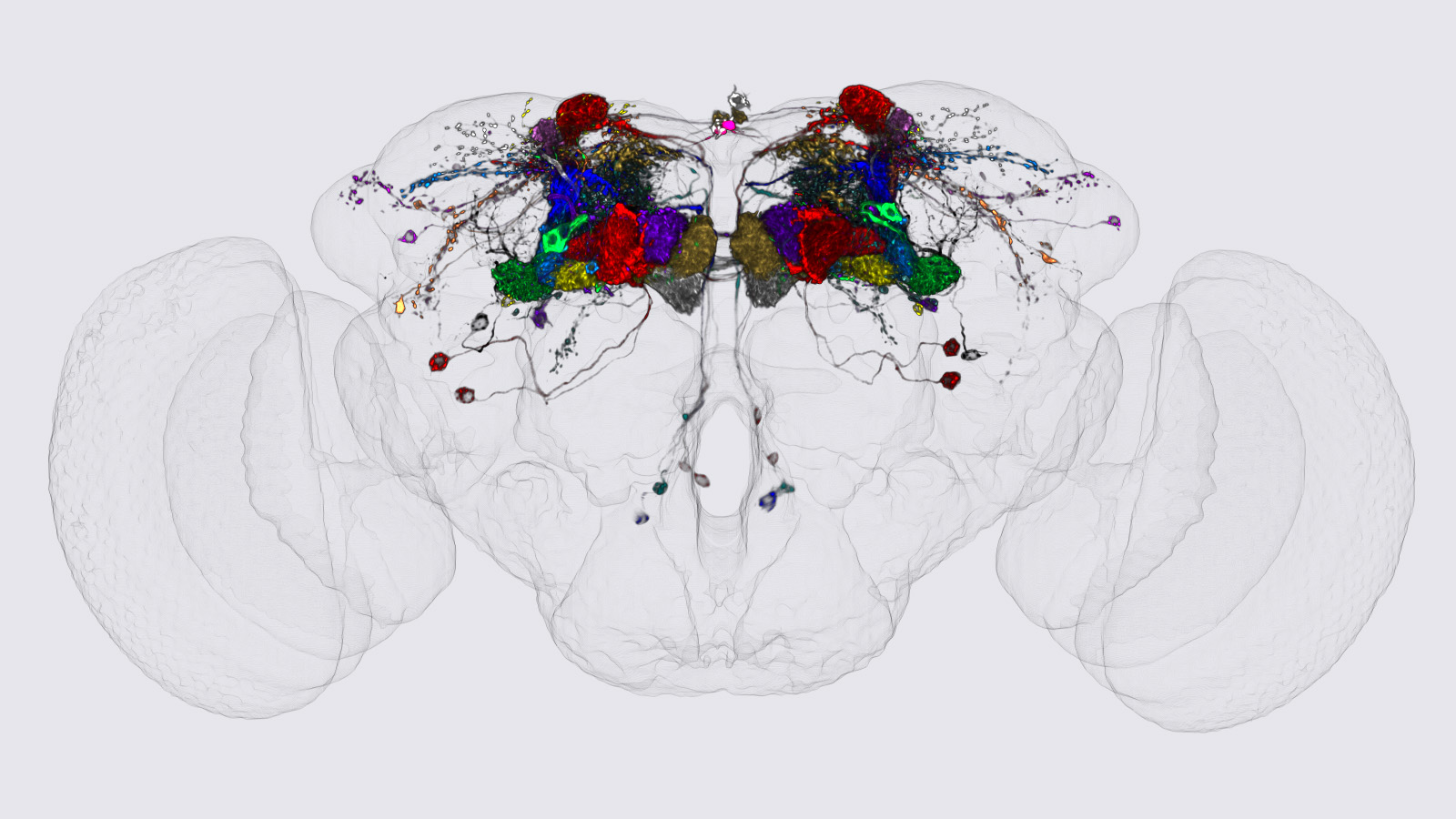
View Publication PageWe identified the neurons comprising the Drosophila mushroom body (MB), an associative center in invertebrate brains, and provide a comprehensive map describing their potential connections. Each of the 21 MB output neuron (MBON) types elaborates segregated dendritic arbors along the parallel axons of ∼2000 Kenyon cells, forming 15 compartments that collectively tile the MB lobes. MBON axons project to five discrete neuropils outside of the MB and three MBON types form a feedforward network in the lobes. Each of the 20 dopaminergic neuron (DAN) types projects axons to one, or at most two, of the MBON compartments. Convergence of DAN axons on compartmentalized Kenyon cell-MBON synapses creates a highly ordered unit that can support learning to impose valence on sensory representations. The elucidation of the complement of neurons of the MB provides a comprehensive anatomical substrate from which one can infer a functional logic of associative olfactory learning and memory.
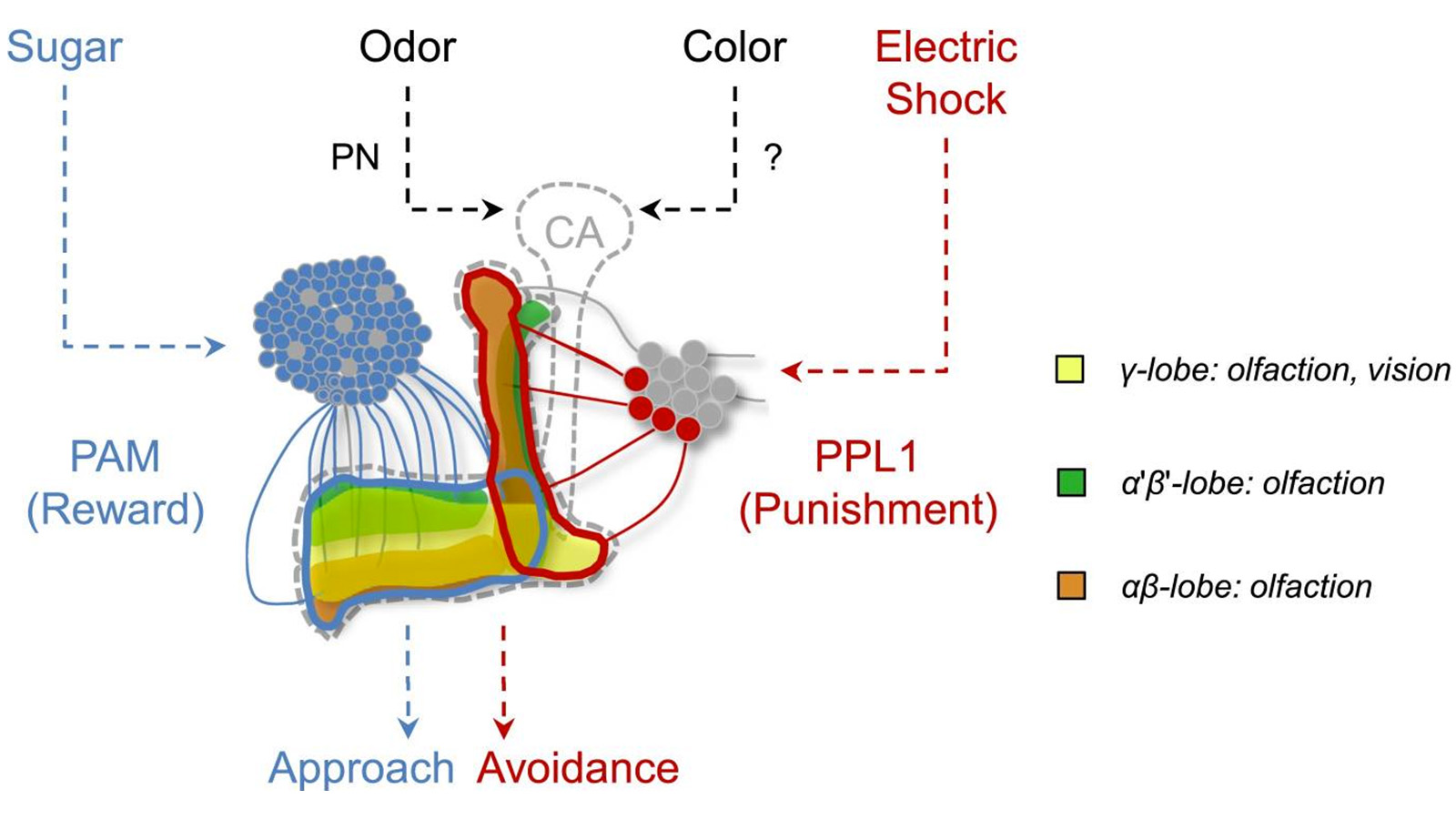
View Publication PageIn nature, animals form memories associating reward or punishment with stimuli from different sensory modalities, such as smells and colors. It is unclear, however, how distinct sensory memories are processed in the brain. We established appetitive and aversive visual learning assays for Drosophila that are comparable to the widely used olfactory learning assays. These assays share critical features, such as reinforcing stimuli (sugar reward and electric shock punishment), and allow direct comparison of the cellular requirements for visual and olfactory memories. We found that the same subsets of dopamine neurons drive formation of both sensory memories. Furthermore, distinct yet partially overlapping subsets of mushroom body intrinsic neurons are required for visual and olfactory memories. Thus, our results suggest that distinct sensory memories are processed in a common brain center. Such centralization of related brain functions is an economical design that avoids the repetition of similar circuit motifs.
View Publication PageAn important strategy for efficient neural coding is to match the range of cellular responses to the distribution of relevant input signals. However, the structure and relevance of sensory signals depend on behavioral state. Here, we show that behavior modifies neural activity at the earliest stages of fly vision. We describe a class of wide-field neurons that provide feedback to the most peripheral layer of the Drosophila visual system, the lamina. Using in vivo patch-clamp electrophysiology, we found that lamina wide-field neurons respond to low-frequency luminance fluctuations. Recordings in flying flies revealed that the gain and frequency tuning of wide-field neurons change during flight, and that these effects are mimicked by the neuromodulator octopamine. Genetically silencing wide-field neurons increased behavioral responses to slow-motion stimuli. Together, these findings identify a cell type that is gated by behavior to enhance neural coding by subtracting low-frequency signals from the inputs to motion detection circuits.
View Publication PageMotion detection is a fundamental neural computation performed by many sensory systems. In the fly, local motion computation is thought to occur within the first two layers of the visual system, the lamina and medulla. We constructed specific genetic driver lines for each of the 12 neuron classes in the lamina. We then depolarized and hyperpolarized each neuron type and quantified fly behavioral responses to a diverse set of motion stimuli. We found that only a small number of lamina output neurons are essential for motion detection, while most neurons serve to sculpt and enhance these feedforward pathways. Two classes of feedback neurons (C2 and C3), and lamina output neurons (L2 and L4), are required for normal detection of directional motion stimuli. Our results reveal a prominent role for feedback and lateral interactions in motion processing and demonstrate that motion-dependent behaviors rely on contributions from nearly all lamina neuron classes.
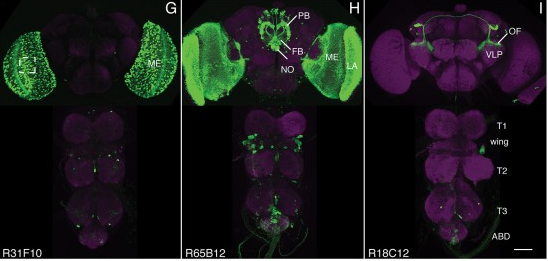
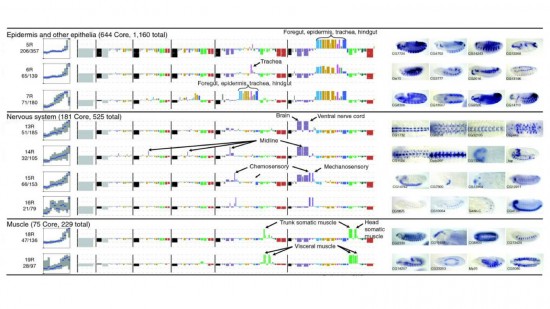
View Publication PageWe established a collection of 7,000 transgenic lines of Drosophila melanogaster. Expression of GAL4 in each line is controlled by a different, defined fragment of genomic DNA that serves as a transcriptional enhancer. We used confocal microscopy of dissected nervous systems to determine the expression patterns driven by each fragment in the adult brain and ventral nerve cord. We present image data on 6,650 lines. Using both manual and machine-assisted annotation, we describe the expression patterns in the most useful lines. We illustrate the utility of these data for identifying novel neuronal cell types, revealing brain asymmetry, and describing the nature and extent of neuronal shape stereotypy. The GAL4 lines allow expression of exogenous genes in distinct, small subsets of the adult nervous system. The set of DNA fragments, each driving a documented expression pattern, will facilitate the generation of additional constructs for manipulating neuronal function. synapse was substantially elevated, at the endocytic zone there was no enhanced polymerization activity. We conclude that actin subserves spatially diverse, independently regulated processes throughout spines. Perisynaptic actin forms a uniquely dynamic structure well suited for direct, active regulation of the synapse.
For the overall strategy and methods used to produce the GAL4 lines:
Pfeiffer, B.D., Jenett, A., Hammonds, A.S., Ngo, T.T., Misra, S., Murphy, C., Scully, A., Carlson, J.W., Wan, K.H., Laverty, T.R., Mungall, C., Svirskas, R., Kadonaga, J.T., Doe, C.Q., Eisen, M.B., Celniker, S.E., Rubin, G.M. (2008). Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl. Acad. Sci. USA 105, 9715-9720. http://www.pnas.org/content/105/28/9715.full.pdf+html synapse was substantially elevated, at the endocytic zone there was no enhanced polymerization activity. We conclude that actin subserves spatially diverse, independently regulated processes throughout spines. Perisynaptic actin forms a uniquely dynamic structure well suited for direct, active regulation of the synapse.
For data on expression in the embryo:
Manning, L., Purice, M.D., Roberts, J., Pollard, J.L., Bennett, A.L., Kroll, J.R., Dyukareva, A.V., Doan, P.N., Lupton, J.R., Strader, M.E., Tanner, S., Bauer, D., Wilbur, A., Tran, K.D., Laverty, T.R., Pearson, J.C., Crews, S.T., Rubin, G.M., and Doe, C.Q. (2012) Annotated embryonic CNS expression patterns of 5000 GMR GAL4 lines: a resource for manipulating gene expression and analyzing cis-regulatory motifs. Cell Reports (2012) Doi: 10.1016/j.celrep.2012.09.009 http://www.cell.com/cell-reports/fulltext/S2211-1247(12)00290-2 synapse was substantially elevated, at the endocytic zone there was no enhanced polymerization activity. We conclude that actin subserves spatially diverse, independently regulated processes throughout spines. Perisynaptic actin forms a uniquely dynamic structure well suited for direct, active regulation of the synapse.
For data on expression in imaginal discs:
Jory, A., Estella, C., Giorgianni, M.W., Slattery, M., Laverty, T.R., Rubin, G.M., and Mann, R.S. (2012) A survey of 6300 genomic fragments for cis-regulatory activity in the imaginal discs of Drosophila melanogaster. Cell Reports (2012) Doi: 10.1016/j.celrep.2012.09.010 http://www.cell.com/cell-reports/fulltext/S2211-1247(12)00291-4 synapse was substantially elevated, at the endocytic zone there was no enhanced polymerization activity. We conclude that actin subserves spatially diverse, independently regulated processes throughout spines. Perisynaptic actin forms a uniquely dynamic structure well suited for direct, active regulation of the synapse.
For data on expression in the larval nervous system:
Li, H.-H., Kroll, J.R., Lennox, S., Ogundeyi, O., Jeter, J., Depasquale, G., and Truman, J.W. (2013) A GAL4 driver resource for developmental and behavioral studies on the larval CNS of Drosophila. Cell Reports (submitted).
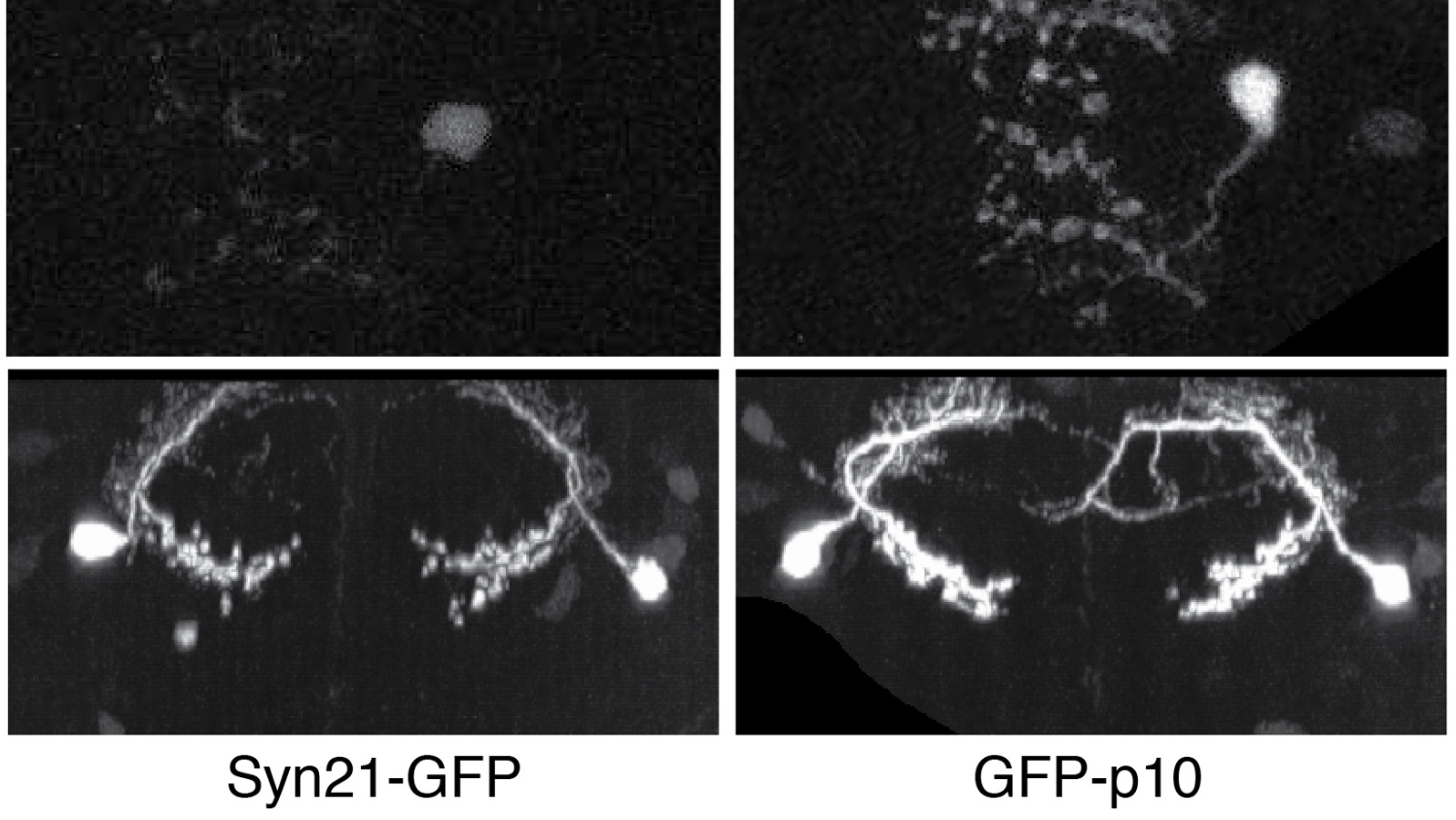
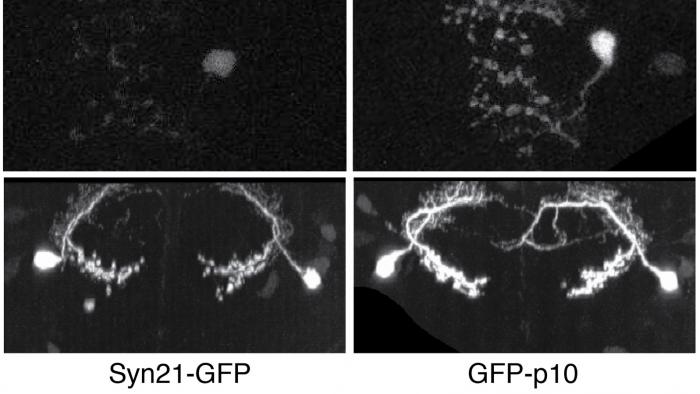
The ability to specify the expression levels of exogenous genes inserted in the genomes of transgenic animals is critical for the success of a wide variety of experimental manipulations. Protein production can be regulated at the level of transcription, mRNA transport, mRNA half-life, or translation efficiency. In this report, we show that several well-characterized sequence elements derived from plant and insect viruses are able to function in Drosophila to increase the apparent translational efficiency of mRNAs by as much as 20-fold. These increases render expression levels sufficient for genetic constructs previously requiring multiple copies to be effective in single copy, including constructs expressing the temperature-sensitive inactivator of neuronal function Shibire(ts1), and for the use of cytoplasmic GFP to image the fine processes of neurons.
View Publication PageSite-specific recombinases have been used for two decades to manipulate the structure of animal genomes in highly predictable ways and have become major research tools. However, the small number of recombinases demonstrated to have distinct specificities, low toxicity, and sufficient activity to drive reactions to completion in animals has been a limitation. In this report we show that four recombinases derived from yeast-KD, B2, B3, and R-are highly active and nontoxic in Drosophila and that KD, B2, B3, and the widely used FLP recombinase have distinct target specificities. We also show that the KD and B3 recombinases are active in mice.
View Publication PageAversive olfactory memory is formed in the mushroom bodies in Drosophila melanogaster. Memory retrieval requires mushroom body output, but the manner in which a memory trace in the mushroom body drives conditioned avoidance of a learned odor remains unknown. To identify neurons that are involved in olfactory memory retrieval, we performed an anatomical and functional screen of defined sets of mushroom body output neurons. We found that MB-V2 neurons were essential for retrieval of both short- and long-lasting memory, but not for memory formation or memory consolidation. MB-V2 neurons are cholinergic efferent neurons that project from the mushroom body vertical lobes to the middle superiormedial protocerebrum and the lateral horn. Notably, the odor response of MB-V2 neurons was modified after conditioning. As the lateral horn has been implicated in innate responses to repellent odorants, we propose that MB-V2 neurons recruit the olfactory pathway involved in innate odor avoidance during memory retrieval.
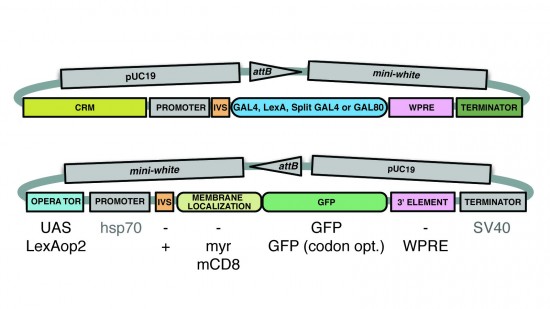
View Publication PageA wide variety of biological experiments rely on the ability to express an exogenous gene in a transgenic animal at a defined level and in a spatially and temporally controlled pattern. We describe major improvements of the methods available for achieving this objective in Drosophila melanogaster. We have systematically varied core promoters, UTRs, operator sequences, and transcriptional activating domains used to direct gene expression with the GAL4, LexA, and Split GAL4 transcription factors and the GAL80 transcriptional repressor. The use of site-specific integration allowed us to make quantitative comparisons between different constructs inserted at the same genomic location. We also characterized a set of PhiC31 integration sites for their ability to support transgene expression of both drivers and responders in the nervous system. The increased strength and reliability of these optimized reagents overcome many of the previous limitations of these methods and will facilitate genetic manipulations of greater complexity and sophistication.
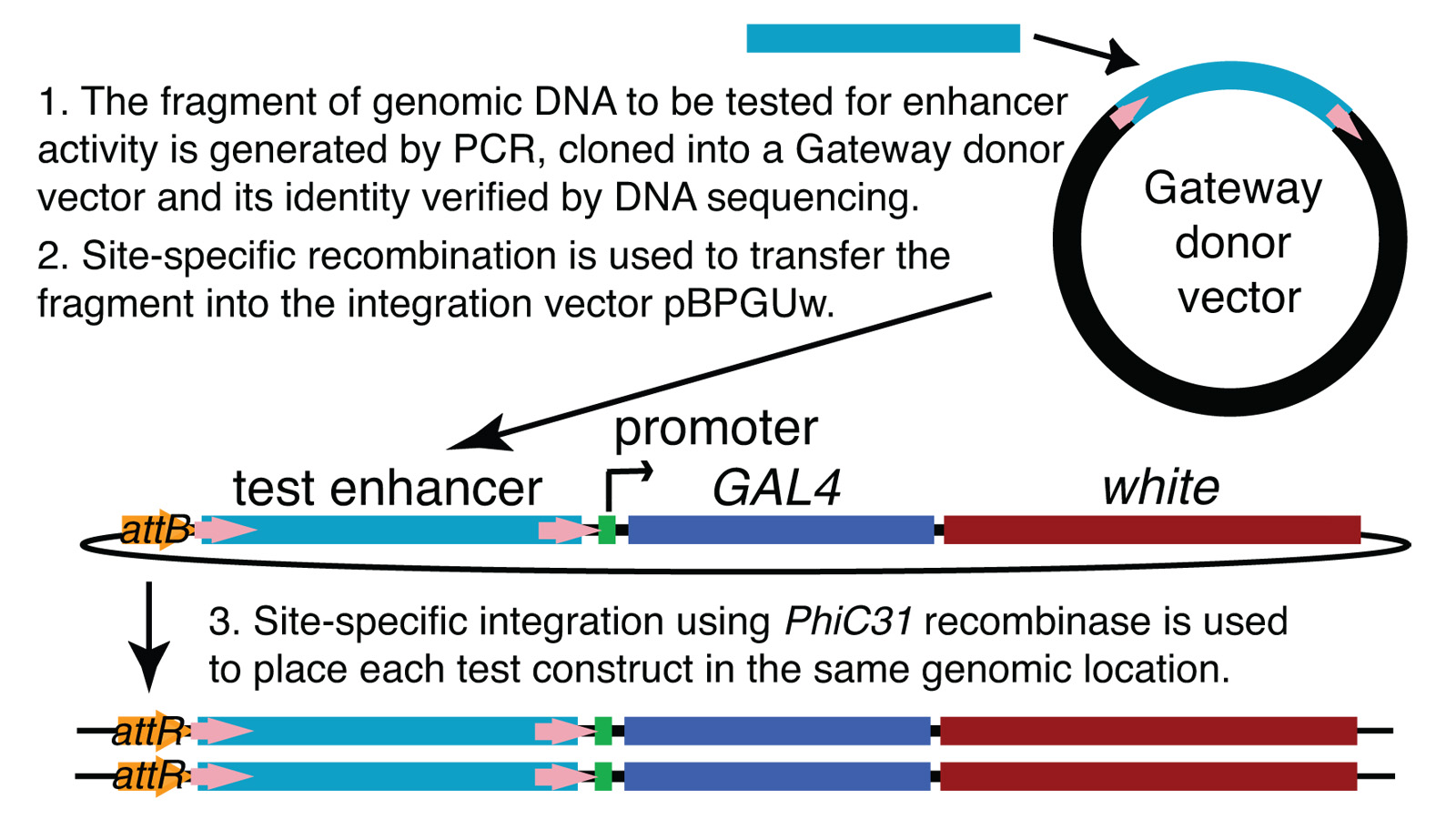
View Publication PageWe demonstrate the feasibility of generating thousands of transgenic Drosophila melanogaster lines in which the expression of an exogenous gene is reproducibly directed to distinct small subsets of cells in the adult brain. We expect the expression patterns produced by the collection of 5,000 lines that we are currently generating to encompass all neurons in the brain in a variety of intersecting patterns. Overlapping 3-kb DNA fragments from the flanking noncoding and intronic regions of genes thought to have patterned expression in the adult brain were inserted into a defined genomic location by site-specific recombination. These fragments were then assayed for their ability to function as transcriptional enhancers in conjunction with a synthetic core promoter designed to work with a wide variety of enhancer types. An analysis of 44 fragments from four genes found that >80% drive expression patterns in the brain; the observed patterns were, on average, comprised of <100 cells. Our results suggest that the D. melanogaster genome contains >50,000 enhancers and that multiple enhancers drive distinct subsets of expression of a gene in each tissue and developmental stage. We expect that these lines will be valuable tools for neuroanatomy as well as for the elucidation of neuronal circuits and information flow in the fly brain.
View Publication PageThe conditional expression of hairpin constructs in Drosophila melanogaster has emerged in recent years as a method of choice in functional genomic studies. To date, upstream activating site-driven RNA interference constructs have been inserted into the genome randomly using P-element-mediated transformation, which can result in false negatives due to variable expression. To avoid this problem, we have developed a transgenic RNA interference vector based on the phiC31 site-specific integration method.
View Publication PageCell and tissue specific gene expression is a defining feature of embryonic development in multi-cellular organisms. However, the range of gene expression patterns, the extent of the correlation of expression with function, and the classes of genes whose spatial expression are tightly regulated have been unclear due to the lack of an unbiased, genome-wide survey of gene expression patterns.
View Publication PageJanelia Farm, the new research campus of the Howard Hughes Medical Institute, is an ongoing experiment in the social engineering of research communities.
View Publication PageTranscriptional regulation in eukaryotes generally operates at the level of individual genes. Regulation of sets of adjacent genes by mechanisms operating at the level of chromosomal domains has been demonstrated in a number of cases, but the fraction of genes in the genome subject to regulation at this level is unknown.
View Publication PageA comparative analysis of the genomes of Drosophila melanogaster, Caenorhabditis elegans, and Saccharomyces cerevisiae-and the proteins they are predicted to encode-was undertaken in the context of cellular, developmental, and evolutionary processes. The nonredundant protein sets of flies and worms are similar in size and are only twice that of yeast, but different gene families are expanded in each genome, and the multidomain proteins and signaling pathways of the fly and worm are far more complex than those of yeast. The fly has orthologs to 177 of the 289 human disease genes examined and provides the foundation for rapid analysis of some of the basic processes involved in human disease.
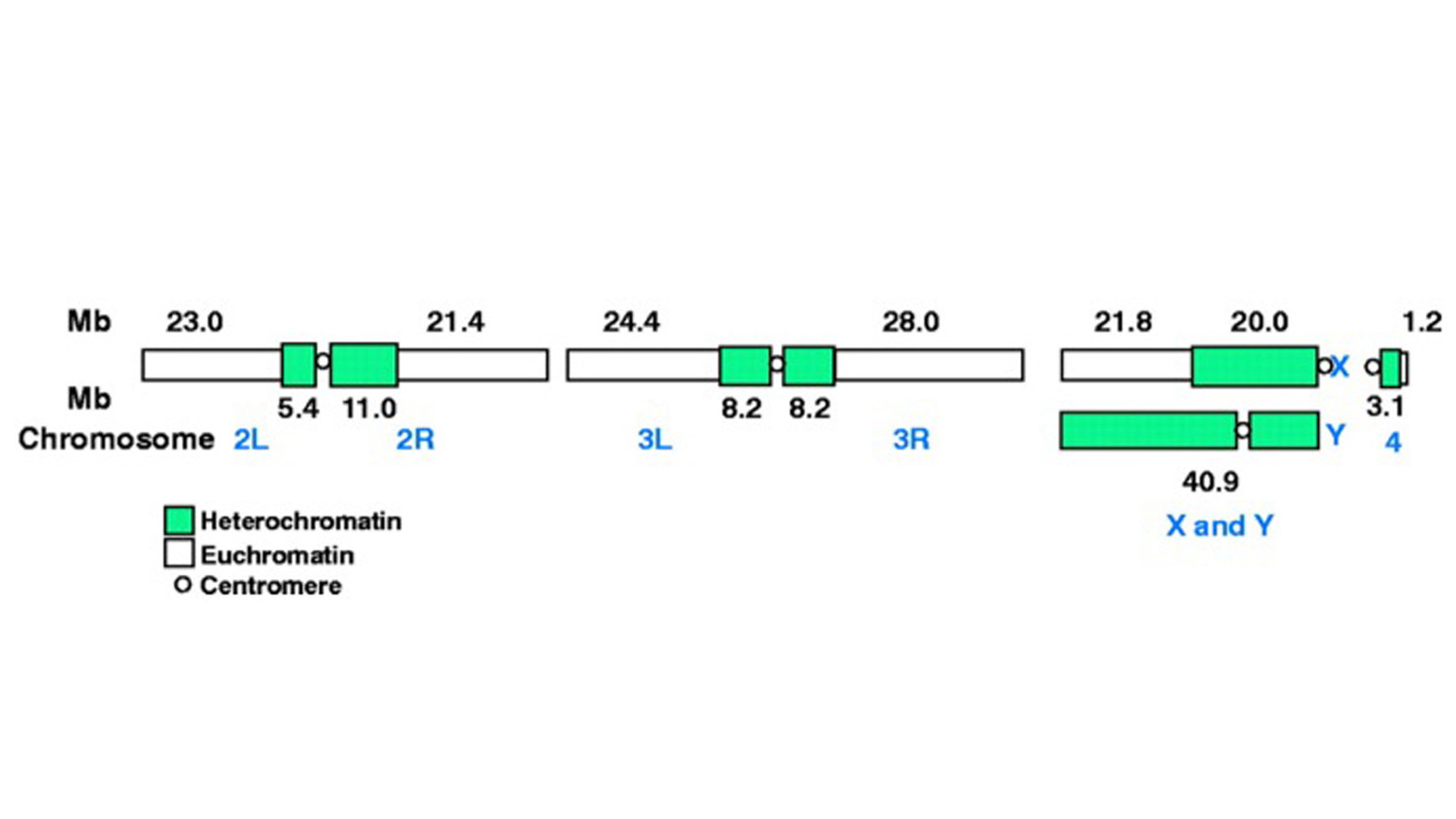
View Publication PageThe fly Drosophila melanogaster is one of the most intensively studied organisms in biology and serves as a model system for the investigation of many developmental and cellular processes common to higher eukaryotes, including humans. We have determined the nucleotide sequence of nearly all of the approximately 120-megabase euchromatic portion of the Drosophila genome using a whole-genome shotgun sequencing strategy supported by extensive clone-based sequence and a high-quality bacterial artificial chromosome physical map. Efforts are under way to close the remaining gaps; however, the sequence is of sufficient accuracy and contiguity to be declared substantially complete and to support an initial analysis of genome structure and preliminary gene annotation and interpretation. The genome encodes approximately 13,600 genes, somewhat fewer than the smaller Caenorhabditis elegans genome, but with comparable functional diversity.
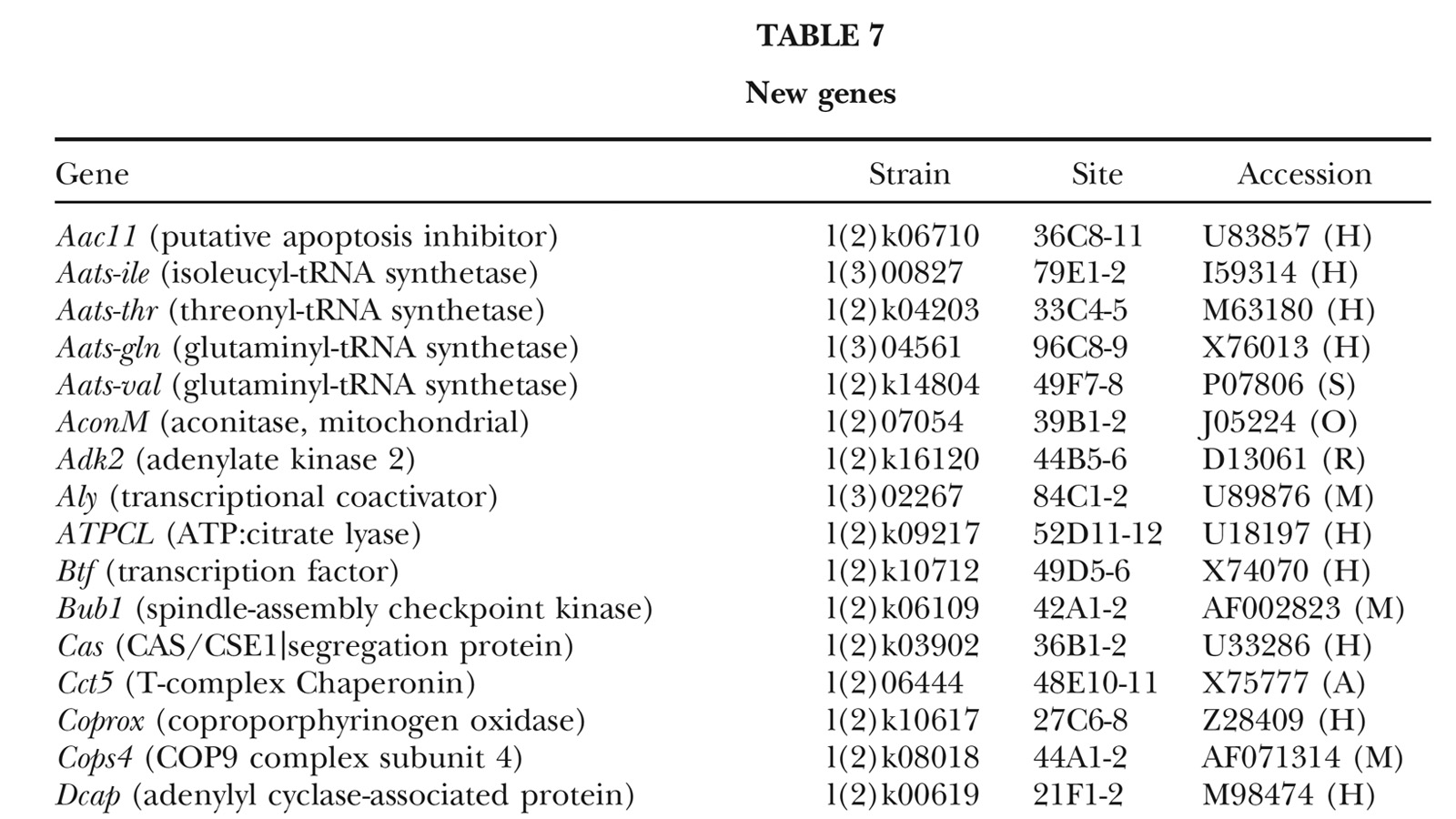
View Publication PageA fundamental goal of genetics and functional genomics is to identify and mutate every gene in model organisms such as Drosophila melanogaster. The Berkeley Drosophila Genome Project (BDGP) gene disruption project generates single P-element insertion strains that each mutate unique genomic open reading frames. Such strains strongly facilitate further genetic and molecular studies of the disrupted loci, but it has remained unclear if P elements can be used to mutate all Drosophila genes. We now report that the primary collection has grown to contain 1045 strains that disrupt more than 25% of the estimated 3600 Drosophila genes that are essential for adult viability. Of these P insertions, 67% have been verified by genetic tests to cause the associated recessive mutant phenotypes, and the validity of most of the remaining lines is predicted on statistical grounds. Sequences flanking >920 insertions have been determined to exactly position them in the genome and to identify 376 potentially affected transcripts from collections of EST sequences. Strains in the BDGP collection are available from the Bloomington Stock Center and have already assisted the research community in characterizing >250 Drosophila genes. The likely identity of 131 additional genes in the collection is reported here. Our results show that Drosophila genes have a wide range of sensitivity to inactivation by P elements, and provide a rationale for greatly expanding the BDGP primary collection based entirely on insertion site sequencing. We predict that this approach can bring >85% of all Drosophila open reading frames under experimental control.
View Publication PageApoptotic cell death is a mechanism by which organisms eliminate superfluous or harmful cells. Expression of the cell death regulatory protein REAPER (RPR) in the developing Drosophila eye results in a small eye owing to excess cell death. We show that mutations in thread (th) are dominant enhancers of RPR-induced cell death and that th encodes a protein homologous to baculovirus inhibitors of apoptosis (IAPs), which we call Drosophila IAP1 (DIAP1). Overexpression of DIAP1 or a related protein, DIAP2, in the eye suppresses normally occurring cell death as well as death due to overexpression of rpr or head involution defective. IAP death-preventing activity localizes to the N-terminal baculovirus IAP repeats, a motif found in both viral and cellular proteins associated with death prevention.
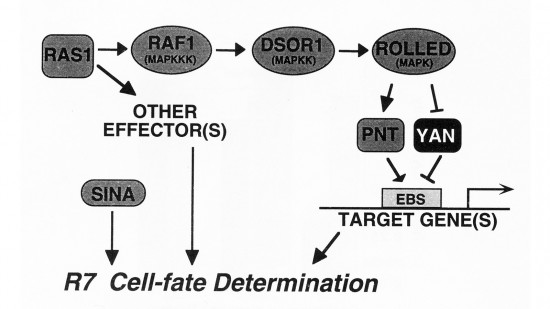
View Publication PageDrosophila yan has been postulated to act as an antagonist of the proneural signal mediated by the sevenless/Ras1/MAPK pathway. We have mutagenized the eight MAPK phosphorylation consensus sites of yan and examined the effects of overexpressing the mutant protein in transgenic flies and transfected S2 cultured cells. Our results suggest that phosphorylation by MAPK affects the stability and subcellular localization of yan, resulting in rapid down-regulation of yan activity. Furthermore, MAPK-mediated down-regulation of yan function appears to be critical for the proper differentiation of both neuronal and nonneuronal tissues throughout development, suggesting that yan is an essential component of a general timing mechanism controlling the competence of a cell to respond to inductive signals.
View Publication PageWe show that the activities of two Ets-related transcription factors required for normal eye development in Drosophila, pointed and yan, are regulated by the Ras1/MAPK pathway. The pointed gene codes for two related proteins, and we show that one form is a constitutive activator of transcription, while the activity of the other form is stimulated by the Ras1/MAPK pathway. Mutation of the single consensus MAPK phosphorylation site in the second form abrogates this responsiveness. yan is a negative regulator of photoreceptor determination, and genetic data suggest that it acts as an antagonist of Ras1. We demonstrate that yan can repress transcription and that this repression activity is negatively regulated by the Ras1/MAPK signal, most likely through direct phosphorylation of yan by MAPK.
View Publication PageWe have identified a Drosophila gene, peanut (pnut), that is related in sequence to the CDC3, CDC10, CDC11, and CDC12 genes of S. cerevisiae. These genes are required for cytokinesis, and their products are present at the bud neck during cell division. We find that pnut is also required for cytokinesis: in pnut mutants, imaginal tissues fail to proliferate and instead develop clusters of large, multinucleate cells. Pnut protein is localized to the cleavage furrow of dividing cells during cytokinesis and to the intercellular bridge connecting postmitotic daughter cells. In addition to its role in cytokinesis, pnut displays genetic interactions with seven in absentia, a gene required for neuronal fate determination in the compound eye, suggesting that pnut may have pleiotropic functions. Our results suggest that this class of proteins is involved in aspects of cytokinesis that have been conserved between flies and yeast.
View Publication PageDevelopment of the Drosophila retina occurs asynchronously; differentiation, its front marked by the morphogenetic furrow, progresses across the eye disc epithelium over a 2 day period. We have investigated the mechanism by which this front advances, and our results suggest that developing retinal cells drive the progression of morphogenesis utilizing the products of the hedgehog (hh) and decapentaplegic (dpp) genes. Analysis of hh and dpp genetic mosaics indicates that the products of these genes act as diffusible signals in this process. Expression of dpp in the morphogenetic furrow is closely correlated with the progression of the furrow under a variety of conditions. We show that hh, synthesized by differentiating cells, induces the expression of dpp, which appears to be a primary mediator of furrow movement.
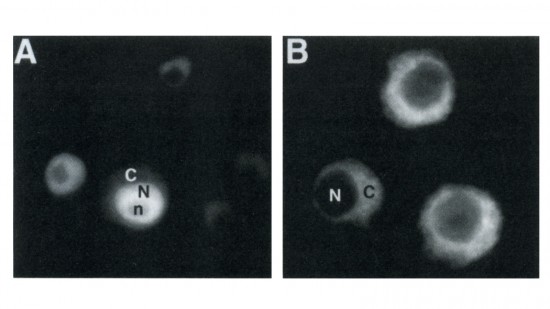
View Publication PageWe have constructed a series of strains to facilitate the generation and analysis of clones of genetically distinct cells in developing and adult tissues of Drosophila. Each of these strains carries an FRT element, the target for the yeast FLP recombinase, near the base of a major chromosome arm, as well as a gratuitous cell-autonomous marker. Novel markers that carry epitope tags and that are localized to either the cell nucleus or cell membrane have been generated. As a demonstration of how these strains can be used to study a particular gene, we have analyzed the developmental role of the Drosophila EGF receptor homolog. Moreover, we have shown that these strains can be utilized to identify new mutations in mosaic animals in an efficient and unbiased way, thereby providing an unprecedented opportunity to perform systematic genetic screens for mutations affecting many biological processes.
View Publication PageThe argos gene encodes a protein that is required for viability and that regulates the determination of cells in the Drosophila eye. A developmental analysis of argos mutant eyes indicates that the mystery cells, which are usually nonneuronal, are transformed into extra photoreceptors, and that supernumerary cone cells and pigment cells are also recruited. Clonal analysis indicates that argos acts nonautonomously and can diffuse over the range of several cell diameters. Conceptual translation of the argos gene suggests that it encodes a secreted protein.
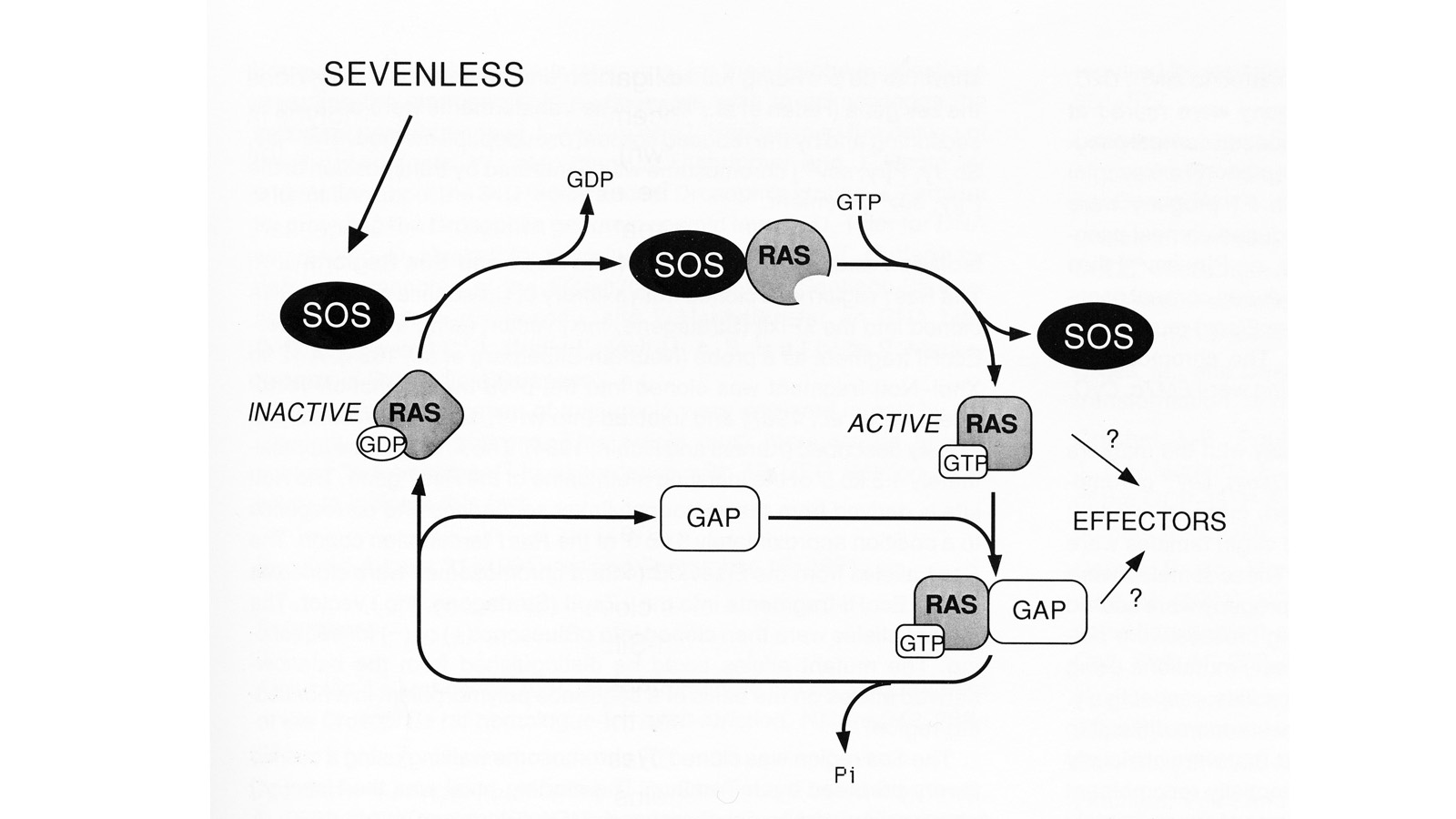
View Publication PageWe have conducted a genetic screen for mutations that decrease the effectiveness of signaling by a protein tyrosine kinase, the product of the Drosophila melanogaster sevenless gene. These mutations define seven genes whose wild-type products may be required for signaling by sevenless. Four of the seven genes also appear to be essential for signaling by a second protein tyrosine kinase, the product of the Ellipse gene. The putative products of two of these seven genes have been identified. One encodes a ras protein. The other locus encodes a protein that is homologous to the S. cerevisiae CDC25 protein, an activator of guanine nucleotide exchange by ras proteins. These results suggest that the stimulation of ras protein activity is a key element in the signaling by sevenless and Ellipse and that this stimulation may be achieved by activating the exchange of GTP for bound GDP by the ras protein.
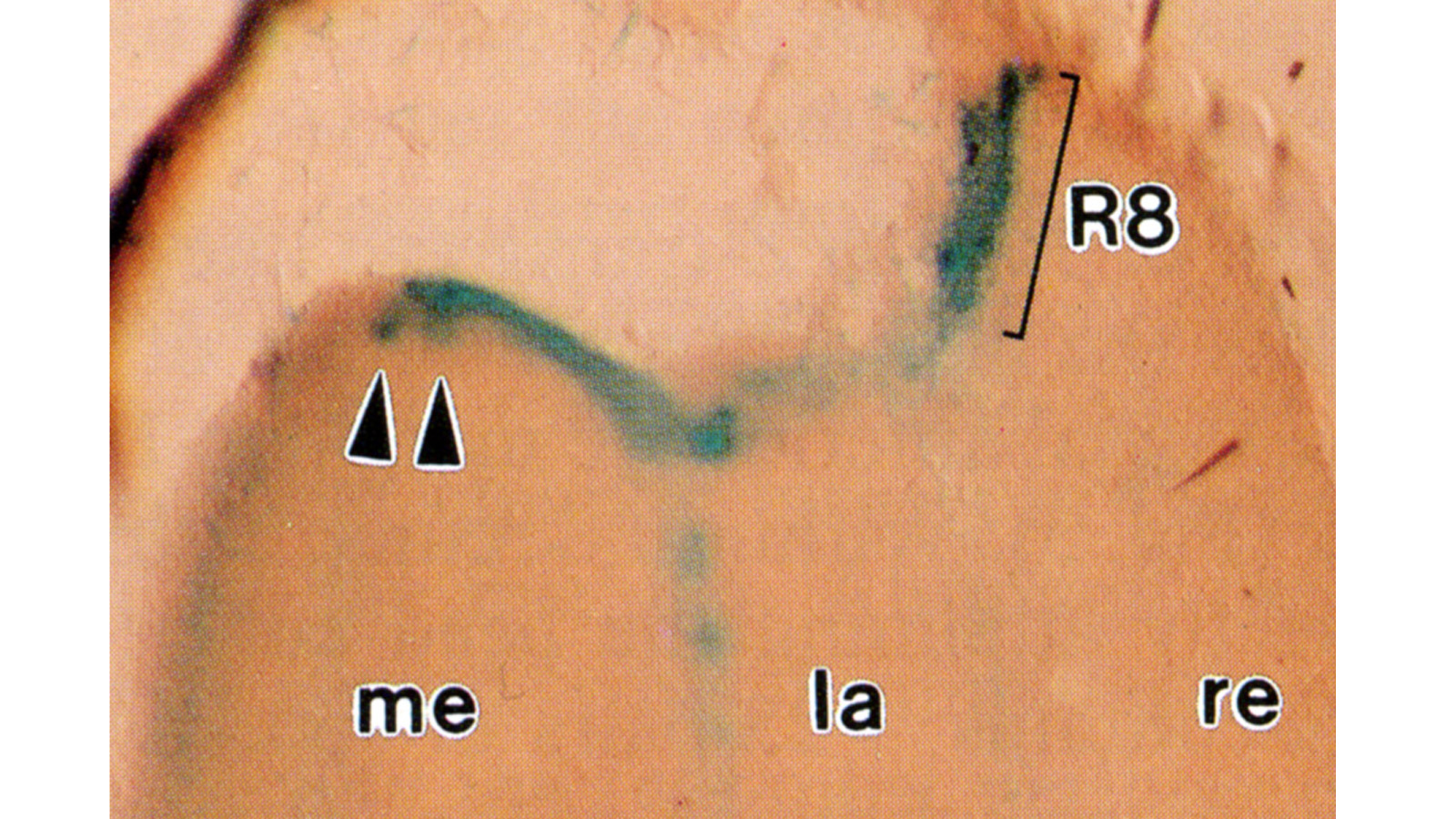
View Publication PageHistological staining of wild-type and sevenless transgenic Drosophila melanogaster bearing Rh3-lacZ fusion genes permits the selective visualization of polarization-sensitive R7 and R8 photoreceptor cells located along the dorsal anterior eye margin. Diffusion of beta-galactosidase throughout these cells reveals that they project long axons to the two most peripheral synaptic target rows of the dorsal posterior medulla, defining a specialized marginal zone of this optic lobe. Comparison of the staining patterns of marginal and nonmarginal Rh3-lacZ-expressing photoreceptor cells in the same histological preparations suggest that the marginal cells possess morphologically specialized axons and synaptic terminals. These findings are discussed with reference to the neuroanatomy of the corresponding dorsal marginal eye and optic lobe regions of the larger dipterans Musca and Calliphora, and in relation to the ability of Drosophila to orient to polarized light.
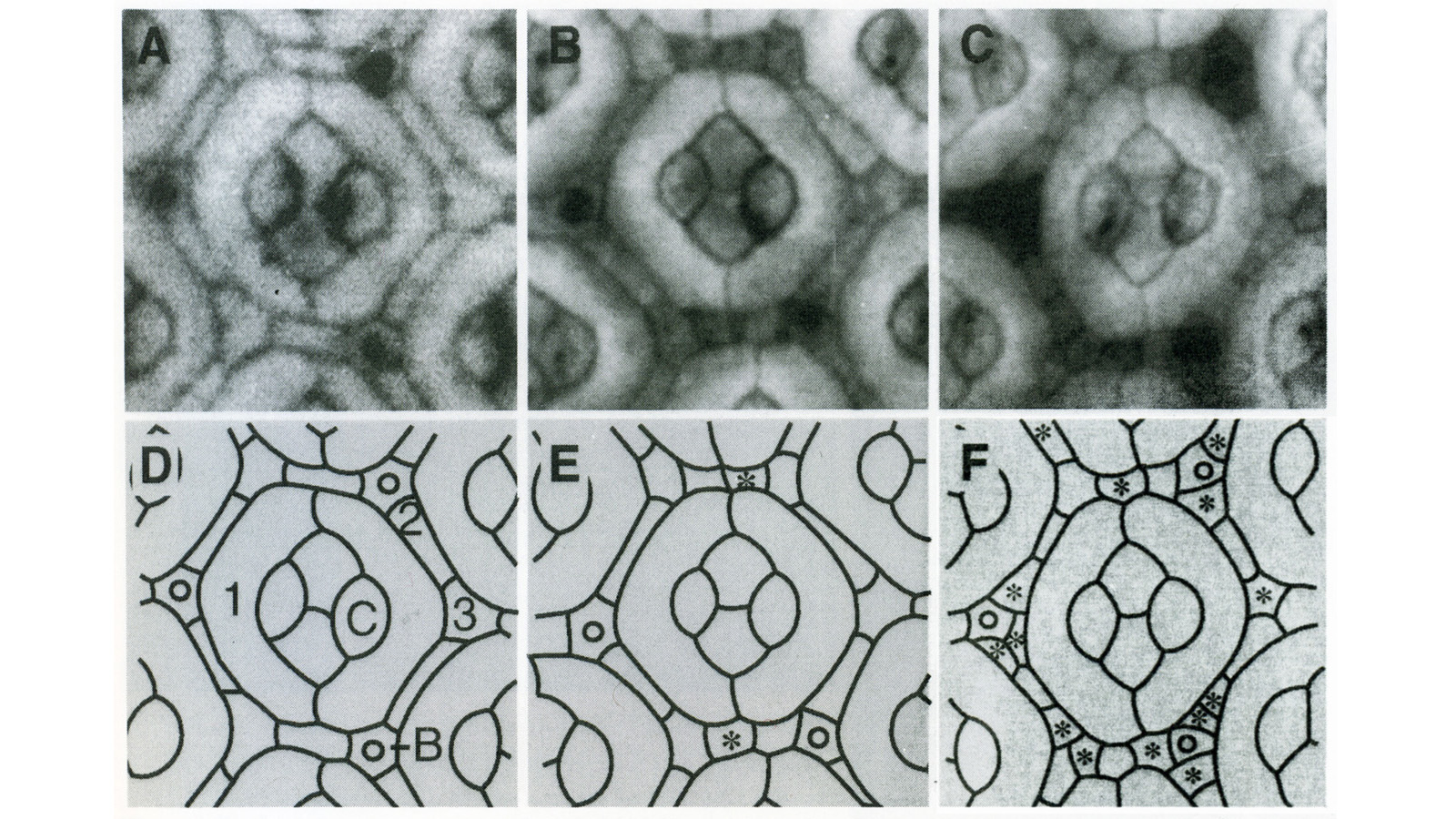
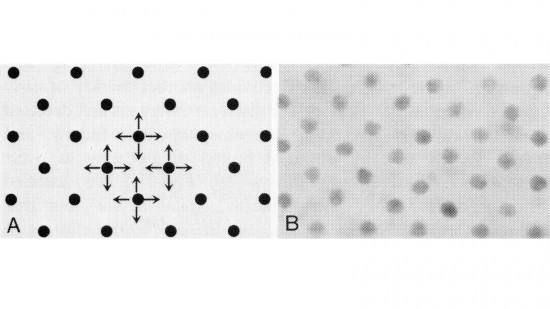
View Publication PageIn the development of multicellular organisms a diversity of cell types differentiate at specific positions. Spacing patterns, in which an array of two or more cell types forms from a uniform field of cells, are a common feature of development. Identical precursor cells may adopt different fates because of competition and inhibition between them. Such a pattern in the developing Drosophila eye is the evenly spaced array of R8 cells, around which other cell types are subsequently recruited. Genetic studies suggest that the scabrous mutation disrupts a signal produced by R8 cells that inhibits other cells from also becoming R8 cells. The scabrous locus was cloned, and it appears to encode a secreted protein partly related to the beta and gamma chains of fibrinogen. It is proposed that the sca locus encodes a lateral inhibitor of R8 differentiation. The roles of the Drosophila EGF-receptor homologue (DER) and Notch genes in this process were also investigated.
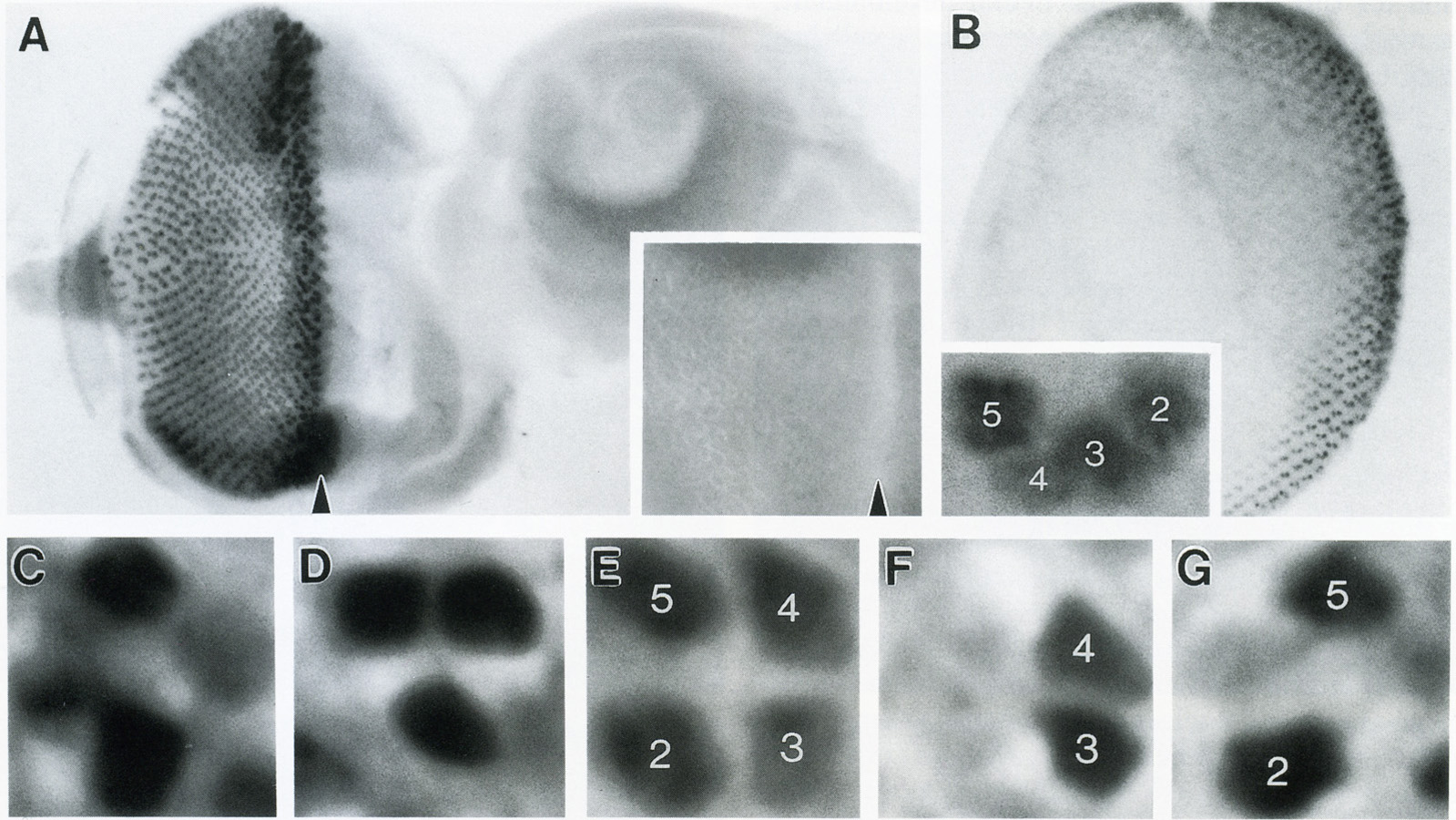
View Publication PageThe Drosophila homeo box gene rough is required in photoreceptor cells R2 and R5 for normal eye development. We show here that rough protein expression is limited to a subset of cells in the developing retina where it is transiently expressed for 30-60 hr. The rough protein is first expressed broadly in the morphogenetic furrow but is rapidly restricted to the R2, R3, R4, and R5 precursor cells. Ubiquitous expression of rough under the control of the hsp70 promoter in third-instar larvae suppresses the initial steps of ommatidial assembly. Structures derived from other imaginal discs are not affected. Ectopic expression of rough in the R7 precursor, through the use of the sevenless promoter, causes this cell to develop into an R1-6 photoreceptor subtype; however, this cell still requires sevenless function for its neural differentiation. Taken together with previous analyses of the rough mutant phenotype, these results suggest that the normal role of rough is to establish the unique cell identity of photoreceptors R2 and R5.
View Publication PageThe rhodopsin genes of Drosophila melanogaster are expressed in nonoverlapping subsets of photoreceptor cells within the insect visual system. Two of these genes, Rh3 and Rh4, are known to display complementary expression patterns in the UV-sensitive R7 photoreceptor cell population of the compound eye. In addition, we find that Rh3 is expressed in a small group of paired R7 and R8 photoreceptor cells at the dorsal eye margin that are apparently specialized for the detection of polarized light. In this paper we present a detailed characterization of the cis-acting requirements of both Rh3 and Rh4. Promoter deletion series demonstrate that small regulatory regions (less than 300 bp) of both R7 opsin genes contain DNA sequences sufficient to generate their respective expression patterns. Individual cis-acting elements were further identified by oligonucleotide-directed mutagenesis guided by interspecific sequence comparisons. Our results suggest that the Drosophila rhodopsin genes share a simple bipartite promoter structure, whereby the proximal region constitutes a functionally equivalent promoter "core" and the distal region determines cell-type specificity. The expression patterns of several hybrid rhodopsin promoters, in which all or part of the putative core regions have been replaced with the analogous regions of different rhodopsin promoters, provide additional evidence in support of this model.
View Publication PageNull mutations of glass specifically remove photoreceptor cells, leaving other cell types intact. We have isolated the glass gene and have shown that its transcript encodes a putative protein of 604 amino acids with five zinc-fingers. The glass product may be a transcription factor required for the development of a single neuronal cell type.
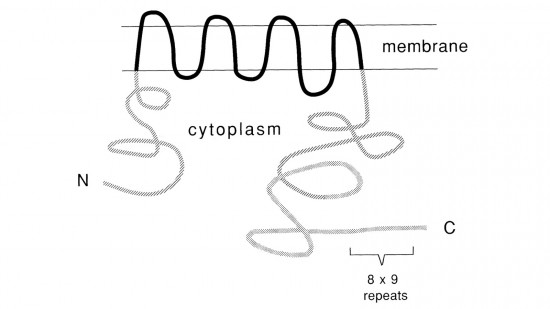
View Publication PageRecent studies suggest that the fly uses the inositol lipid signaling system for visual excitation and that the Drosophila transient receptor potential (trp) mutation disrupts this process subsequent to the production of IP3. In this paper, we show that trp encodes a novel 1275 amino acid protein with eight putative transmembrane segments. Immunolocalization indicates that the trp protein is expressed predominantly in the rhabdomeric membranes of the photoreceptor cells.
View Publication PageThe determination of cell fates during the assembly of the ommatidia in the compound eye of Drosophila appears to be controlled by cell-cell interactions. In this process, the sevenless gene is essential for the development of a single type of photoreceptor cell. In the absence of proper sevenless function the cells that would normally become the R7 photoreceptors instead become nonneuronal cells. Previous morphological and genetic analysis has indicated that the product of the sevenless gene is involved in reading or interpreting the positional information that specifies this particular developmental pathway. The sevenless gene has now been isolated and characterized. The data indicate that sevenless encodes a transmembrane protein with a tyrosine kinase domain. This structural similarity between sevenless and certain hormone receptors suggests that similar mechanisms are involved in developmental decisions based on cell-cell interaction and physiological or developmental changes induced by diffusible factors.
View Publication PageWe show that the germline specificity of P element transposition is controlled at the level of mRNA splicing and not at the level of transcription. In the major P element RNA transcript, isolated from somatic cells, the first three open reading frames are joined by the removal of two introns. Using in vitro mutagenesis and genetic analysis we demonstrate the existence of a third intron whose removal is required for transposase production. We propose that this intron is only removed in the germline and that its removal is the sole basis for the germline restriction of P element transposition.
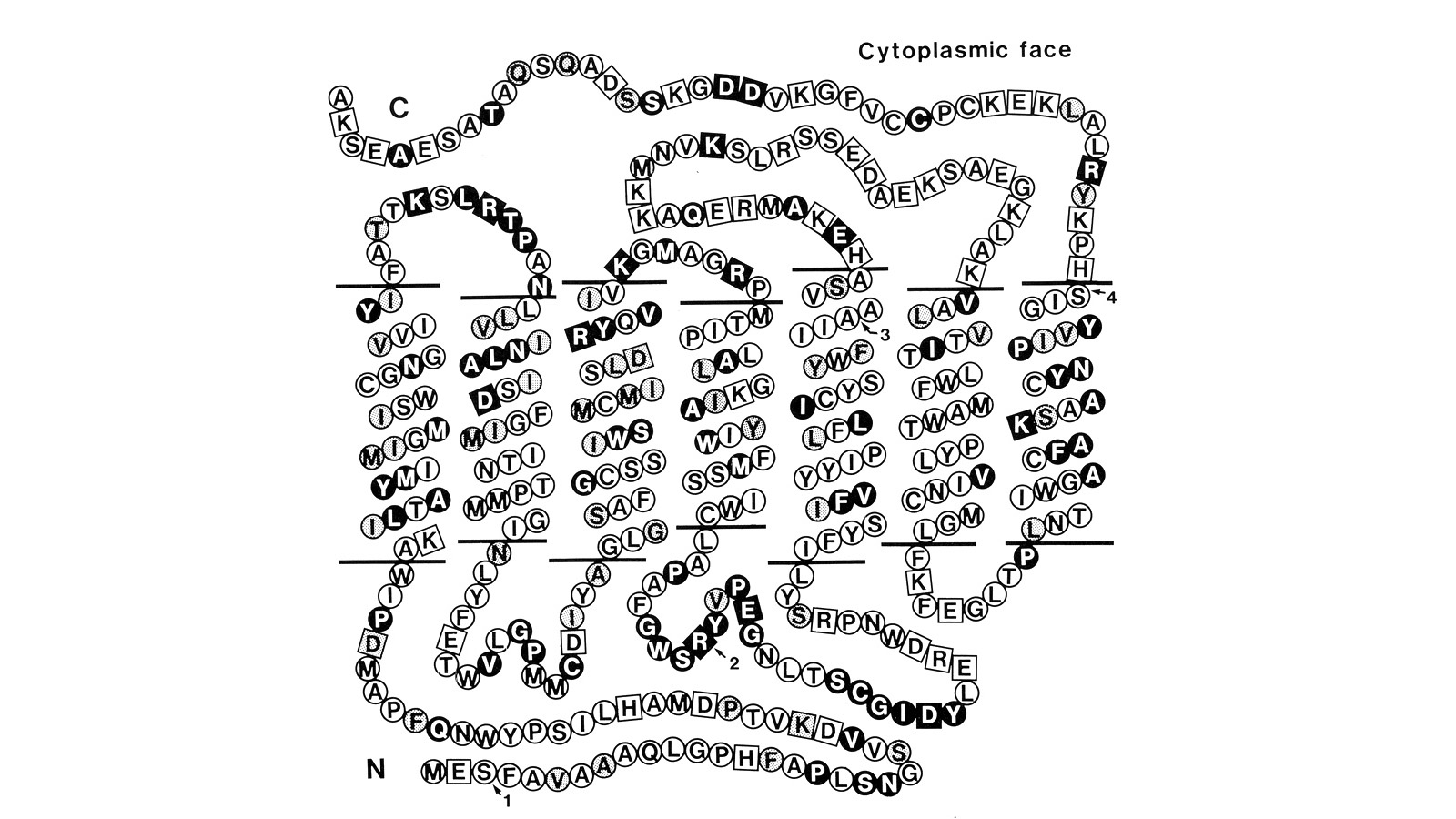
View Publication PageUsing a novel method for detecting cross-homologous nucleic acid sequences we have isolated the gene coding for the major rhodopsin of Drosophila melanogaster and mapped it to chromosomal region 92B8-11. Comparison of cDNA and genomic DNA sequences indicates that the gene is divided into five exons. The amino acid sequence deduced from the nucleotide sequence is 373 residues long, and the polypeptide chain contains seven hydrophobic segments that appear to correspond to the seven transmembrane segments characteristic of other rhodopsins. Three regions of Drosophila rhodopsin are highly conserved with the corresponding domains of bovine rhodopsin, suggesting an important role for these polypeptide regions.
View Publication PageWe have made a P-element derivative called Pc[ry], which carries the selectable marker gene rosy, but which acts like a nondefective, intact P element. It transposes autonomously into the germline chromosomes of an M-strain Drosophila embryo and it mobilizes in trans the defective P elements of the singed-weak allele. Frameshift mutations introduced into any of the four major open reading frames of the P sequence were each sufficient to eliminate the transposase activity, but none affected signals required in cis for transposition of the element. Complementation tests between pairs of mutant elements suggest that a single polypeptide comprises the transposase. We have examined transcripts of P elements both from natural P strains and from lines containing only nondefective Pc[ry] elements, and have identified two RNA species that appear to be specific for autonomous elements.
View Publication PageThirty-six isogenic D. melanogaster strains that differed only in the chromosomal location of a 7.2 or an 8.1 kb DNA segment containing the (autosomal) rosy gene were constructed by P-element-mediated gene transfer. Since the flies were homozygous for a rosy- allele, rosy gene function in these indicated the influence of flanking sequences on gene expression. The tissue distribution of XDH activity in all the strains was normal. Each line exhibited a characteristic level of adult XDH-specific activity. The majority of these values were close to wild-type levels; however, the total variation in specific activity among the lines was nearly fivefold. Thus position effects influence expression of the rosy gene quantitatively but do not detectably alter tissue specificity. X-linked rosy insertions were expressed on average 1.6 times more activity in males than in females. Hence the gene acquires at least partial dosage compensation upon insertion into the X chromosome.
View Publication PageExogenous DNA sequences were introduced into the Drosophila germ line. A rosy transposon (ry1), constructed by inserting a chromosomal DNA fragment containing the wild-type rosy gene into a P transposable element, transformed germ line cells in 20 to 50 percent of the injected rosy mutant embryos. Transformants contained one or two copies of chromosomally integrated, intact ry1 that were stably inherited in subsequent generations. These transformed flies had wild-type eye color indicating that the visible genetic defect in the host strain could be fully and permanently corrected by the transferred gene. To demonstrate the generality of this approach, a DNA segment that does not confer a recognizable phenotype on recipients was also transferred into germ line chromosomes.
View Publication PageWe have shown previously that four of five white mutant alleles arising in P-M dysgenic hybrids result from the insertion of strongly homologous DNA sequence elements. We have named these P elements. We report that P elements are present in 30-50 copies per haploid genome in all P strains examined and apparently are missing entirely from all M strains examined, with one exception. Furthermore, members of the P family apparently transpose frequently in P-M dysgenic hybrids; chromosomes descendant from P-M dysgenic hybrids frequently show newly acquired P elements. Finally, the strain-specific breakpoint hotspots for the rearrangement of the pi 2 P X chromosome occurring in P-M dysgenic hybrids are apparently sites of residence of P elements. These observations strongly support the P factor hypothesis for the mechanistic basis of P-M hybrid dysgenesis.
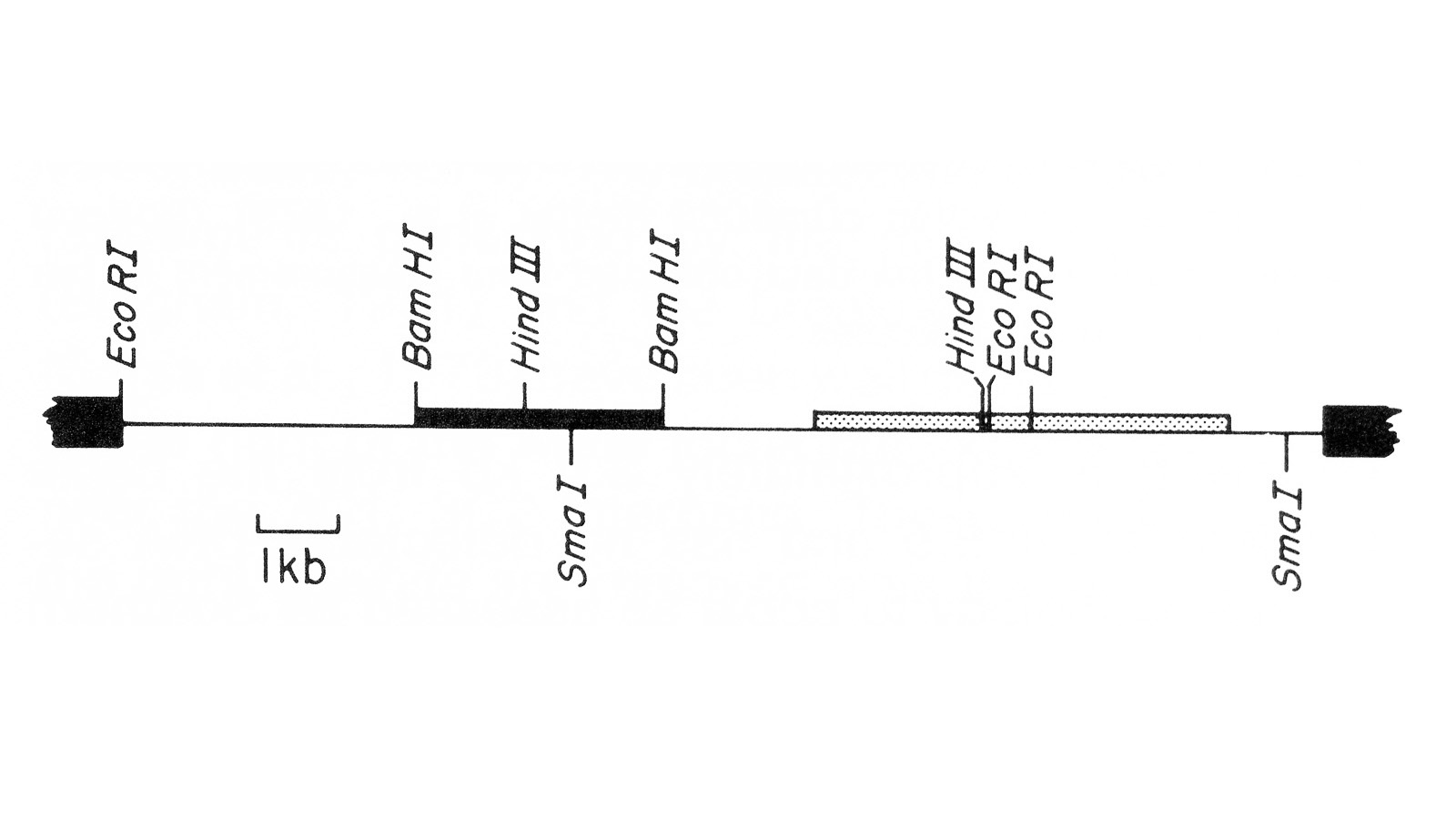
View Publication PageWe describe the isolation of a cloned DNA segment carrying unique sequences from the white locus of Drosophila melanogaster. Sequences within the cloned segment are shown to hybridize in situ to the white locus region on the polytene chromosomes of both wild-type strains and strains carrying chromosomal rearrangements whose breakpoints bracket the white locus. We further show that two small deficiency mutations, deleting white locus genetic elements but not those of complementation groups contiguous to white, delete the genomic sequences corresponding to a portion of the cloned segment. The strategy we have employed to isolate this cloned segment exploits the existence of an allele at the white locus containing a copy of a previously cloned transposable, reiterated DNA sequence element. We describe a simple, rapid method for retrieving cloned segments carrying a copy of the transposable element together with contiguous sequences corresponding to this allele. The strategy described is potentially general and we discuss its application to the cloning of the DNA sequences of other genes in Drosophila, including those identified only by genetic analysis and for which no RNA product is known.
View Publication PageThe stability of elements of three different dispersed repeated gene families in the genome of Drosophila tissue culture cells has been examined. Different amounts of sequences homologous to elements of 412, copia and 297 dispersed repeated gene families are found in the genomes of D. melanogaster embryonic and tissue culture cells. In general the amount of these sequences is increased in the cell lines. The additional sequences homologous to 412, copia and 297 occur as intact elements and are dispersed to new sites in the cell culture genome. It appears that these elements can insert at many alternative sites. We also describe a DNA sequence arrangement found in the D. melanogaster embryo genome which appears to result from a transposition of an element of the copia dispersed repeated gene family into a new chromosomal site. The mechanism of insertion of this copia element is precise to within 90 bp and may involve a region of weak sequence homology between the site of insertion and the direct terminal repeats of the copia element.
View Publication Page