Main Menu (Mobile)- Block
- Overview
-
Support Teams
- Overview
- Anatomy and Histology
- Cryo-Electron Microscopy
- Electron Microscopy
- Flow Cytometry
- Gene Targeting and Transgenics
- High Performance Computing
- Immortalized Cell Line Culture
- Integrative Imaging
- Invertebrate Shared Resource
- Janelia Experimental Technology
- Mass Spectrometry
- Media Prep
- Molecular Genomics
- Primary & iPS Cell Culture
- Project Pipeline Support
- Project Technical Resources
- Quantitative Genomics
- Scientific Computing
- Viral Tools
- Vivarium
- Open Science
- You + Janelia
- About Us
Main Menu - Block
- Overview
- Anatomy and Histology
- Cryo-Electron Microscopy
- Electron Microscopy
- Flow Cytometry
- Gene Targeting and Transgenics
- High Performance Computing
- Immortalized Cell Line Culture
- Integrative Imaging
- Invertebrate Shared Resource
- Janelia Experimental Technology
- Mass Spectrometry
- Media Prep
- Molecular Genomics
- Primary & iPS Cell Culture
- Project Pipeline Support
- Project Technical Resources
- Quantitative Genomics
- Scientific Computing
- Viral Tools
- Vivarium
We build precision imaging tools to probe spatial and temporal control of cell fates during development and in adult tissues.
We are looking for new colleagues. Please visit our external lab website for details: www.liulabspatial.org
A fertilized egg - a single cell - carrying one copy of mom's and one copy of dad's DNA is able to divide and differentiate to trillions of cells and hundreds of cell types in our body, enabling each of us to thrive in the society.
Our lab has focused on developing precision imaging tools to investigate molecular dynamics and cellular structures across various scales related to gene and genome regulation. Our techniques allow for the imaging of nanometer-scale molecular components like proteins and genomic loci in live cells. We also developed super-resolution methods, such as 3D ATAC-PALM, to map genome organization in single cells, helping us study how protein complexes like cohesin regulate chromatin clustering and gene co-expression at the single-cell level.
Inspired by these findings, we recently developed cycleHCR, a method for highly-multiplexed imaging of RNA and proteins in deep tissue context. This tool simultaneously captures spatial gene expression and high-resolution subcellular architecture in thick tissue specimens. Moving forward, our goal is to combine these tools with genetic perturbations to identify networks that regulate high-dimensional gene expression and cell-fate decisions during development and in adult tissues.
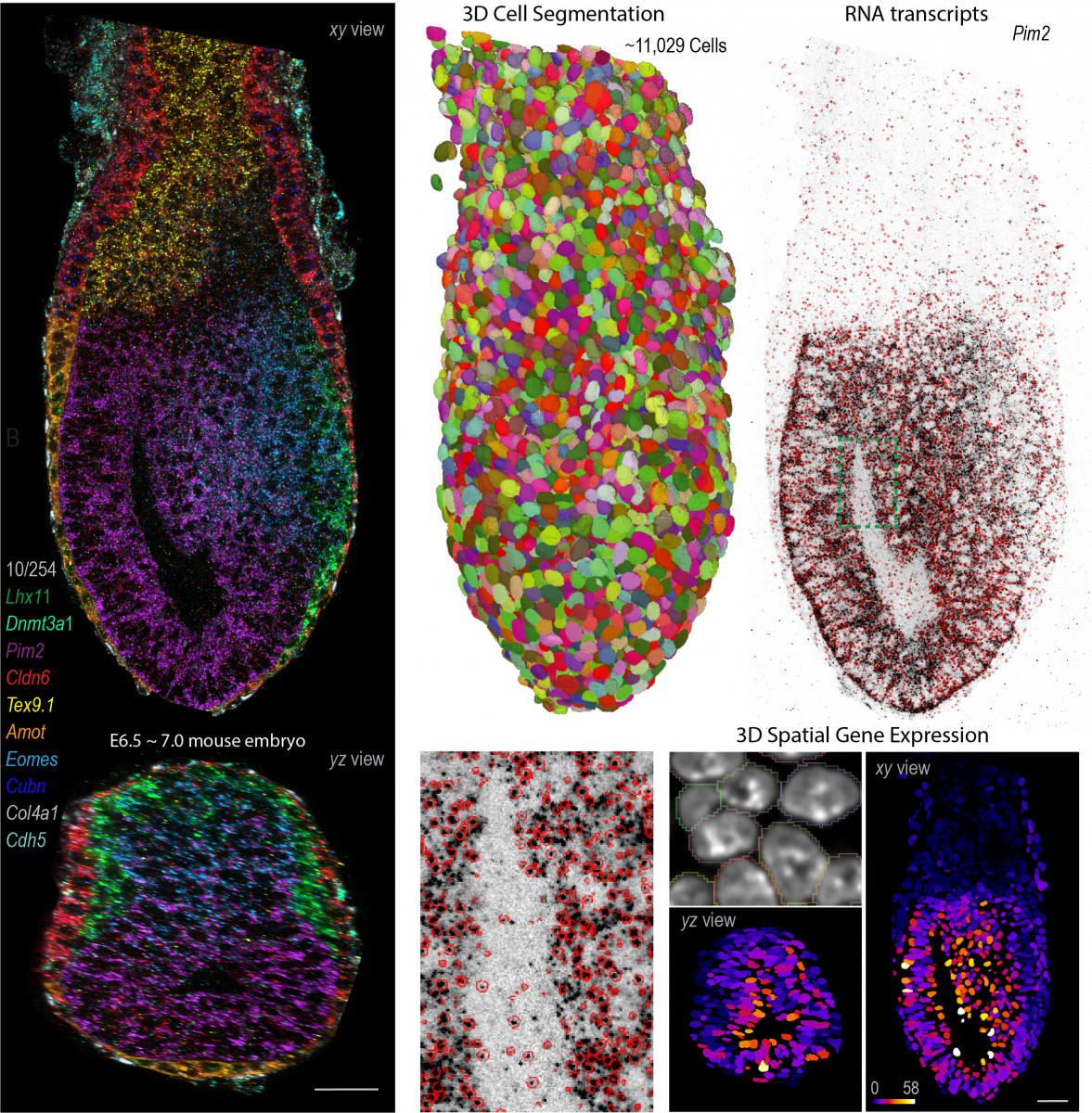
Enabled by the embryo from Alejandro Aguilera Castrejon, Cellpose from Carsen Stringer and RS-FISH from Stephan Preibisch
"Function follows form
at present while form
follows function during
evolution."
- my take
