Sensors for imaging neuronal populations in vivo
The principles of neural control of behavior will emerge from detailed observation and manipulation of neuronal activity. We are engineering fluorescent sensors for imaging of neuronal activity in the intact nervous system of experimental animals. These sensors include calcium indicators, calcium integrators, and voltage indicators. We focus on developing the highest impact sensors that will be useful to the neuroscience community. We screen thousands of variants in neurons and tissue culture cells in vitro and then select the most promising sensors for in vivo characterization.
Specific Aims
-
Produce new GCaMP-type calcium indicators for imaging large neuronal populations and diverse neuron types in fly, mouse, fish, and worms.
-
Engineer red protein calcium indicators with improved sensitivity for dual-color, deep, and channelrhodopsin-compatible imaging.
-
Create transgenic mice expressing indicators of neuronal function for long-term monitoring of neural activity.
-
Optimize genetically-encoded voltage indicators for subcellular voltage imaging and large-scale population imaging.
-
Develop calcium-activated and light-dependent recombinases and transcription factors to couple neural activity and light with gene expression.
Calcium indicators
Understanding the function and plasticity of neural circuits requires measurements of neural activity over a large range of spatial and temporal scales. Calcium indicators have been especially useful for neurobiology. In most neurons, action potentials (APs) are tightly coupled to large (20-fold) and rapid (rise time <1 millisecond) increases in intracellular free calcium concentration. These calcium transients can be measured as fluorescence changes in neuronal somata, dendrites, and axons as a proxy for the underlying electrical activity. Calcium imaging therefore provides a versatile tool to probe neural activity over timescales of milliseconds to months and spatial scales of micrometers to millimeters.
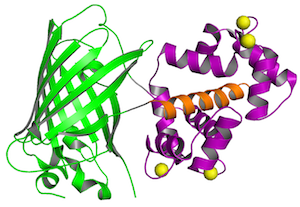
GCaMP detects calcium changes based on a calcium-dependent interaction of calmodulin (purple) and a M13 peptide (orange), which influences the chromophore of a circularly-permuted GFP (green). Four calcium ions (yellow) cooperatively bind to calmodulin. Structure from Akerboom et al., 2009.
Green calcium indicators
The GENIE Project optimizes genetically-encoded calcium indicators (GECIs) to achieve signal-to-noise ratios (SNRs) that rival electrophysiological methods. We have developed the general-purpose GFP-based GCaMP6 sensors, which under favorable conditions detect single spikes in neuronal populations in vivo. GECIs with even higher sensitivity will allow larger populations of neurons to be imaged more rapidly and under more challenging conditions (e.g. during behavior). But high sensitivity comes at the price of limited dynamic range; high-sensitivity indicators saturate at modest activity levels, making them unsuitable for monitoring activity patterns in neurons with high spike rates (~100 Hz). We are developing a suite of green indicators tailored to specific applications, including imaging of large populations of neurons, single-spike detection across neuron types, measurement of spike rates over a wide dynamic range, and imaging spike times with high-speed indicators. We expect to benchmark an additional 500 GCaMP variants.
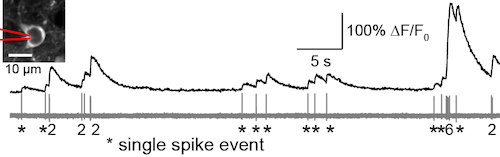
GCaMP6s, an ultrasensitive calcium sensor, detects single spikes with high fidelity in mouse visual cortex (Chen et al., 2013).
Red calcium indicators
Optimized red fluorescent GECIs will enable new in vivo imaging modalities. Longer wavelength excitation light penetrates deeper into tissue. In addition, red fluorescence is absorbed much less than green fluorescence, especially in mammalian tissue, giving red fluorophores a competitive advantage for in vivo imaging. Red and green GECIs can be used for dual color imaging of intermingled neurons. Red GECIs are also more easily combined with channelrhodopsin-2-based manipulations of neural circuits. Channelrhodopsin-2's blue action spectrum is separated from the red GECI excitation spectrum. We have thus developed sensitive red GECIs, including the mApple-based jRGECO1a and mRuby-based jRCaMP1a. After screening 3,000 variants we are currently validating the best red GECIs in vivo. We are in the process of creating viruses, flies and mice expressing these indicators for in vivo imaging.
CaMPARI is a light-gated calcium integrator that permanently marks neurons that were active during user-defined periods. The permanent labeling is based on calcium- and light-dependent photoconversion of EosFP molecules from green to red fluorescence (Fosque et al., 2014).
Calcium integrators and genetic switches
Identifying groups of neurons that are coactive during specific phases of behavior is a major goal of neuroscience. Tools to mark these ensembles in vivo, and to target them for later characterization or manipulation are lacking. Promoters from immediate early genes such as c-fos have been harnessed to trigger transgene expression in active cells, but these methods lack sensitivity, respond with slow kinetics, and are modulated by many physiological parameters other than neural activity. We have developed a photoconverible fluorescent protein, CaMPARI, that can detect the coincidence of light and calcium to mark neurons that are active during experimenter-defined periods. We are developing similar switches to activate gene expression, either through transcriptional regulation of a target gene or through recombination.
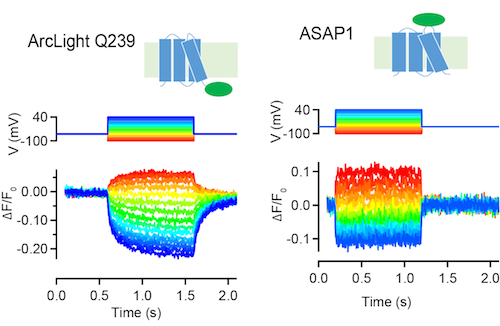
ArcLight Q239 and ASAP1 are GFP-based sensors of membrange voltage (Jin et al., 2012; St-Pierre et al., 2014). These sensors decrease fluorescence in response to depolarizing voltage steps in tissue culture cells (above, S. Brenowitz, unpublished) and neurons.
Voltage indicators
Sensors of membrane voltage, unlike calcium sensors, provide a direct report of neural activity. Electrical measurements report how neuronal input is converted to neuronal output. Voltage imaging in vivo will be useful for detecting both spiking and subthreshold electrical dynamics of individual neurons and neuronal populations. But current protein voltage sensors still suffer from small fluorescence changes in response to APs, low brightness, poor targeting to the plasma membrane, large capacitive load on the membrane, and/or slow response kinetics. We are developing genetically-encoded voltage indicators (GEVIs) for subcellular imaging and for imaging large populations of neurons. We plan to characterize > 100,000 variants of ArcLight, ASAP1, and novel indicators.
Screening and in vivo assays
We develop sensors by coupling structured-guided mutagenesis of proteins with high-throughput, cell-based assays. Lead sensors are then tested in neurons in vivo in fly, mouse, and other organisms.
Neuronal screening platform
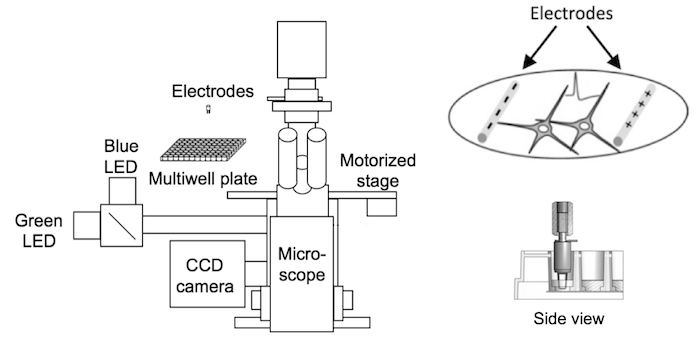
Neurons have fast and small-amplitude calcium accumulations that are difficult to model in non-neuronal systems. For this reason, we developed a cultured neuron screening platform to screen calcium indicators at a rate of thousands per year. Neurons in 96-well plates are transduced with GECI variants. APs are triggered using extracellular stimulation electrodes, while neurons are imaged using fluorescence microscopy. The output of the assay is the fluorescence dynamics of multiple individual neurons to a variety of trains of APs. This assay therefore allows characterization of large numbers of GECI variants under relevant conditions.
The GENIE project's neuronal screening platform combines electrical field stimulation of neurons in multiwell plates, wide-field fluorescence microscopy, and automation. Electrode stimulation (right) evokes action potentials from cultured neurons while sensor fluorescence changes are imaged (Wardill et al., 2013).
GEVI screening platform
We test voltage indicator variants in human embryonic kidney cells expressing voltage-gated sodium and potassium channels. Action potential-like depolarization can be evoked by field stimulation. We leverage the ease of culture and transfection of these cells to achieve high throughput. High-speed imaging is used to capture fast responses to electrical stimuli.
Fly neuromuscular junction assay
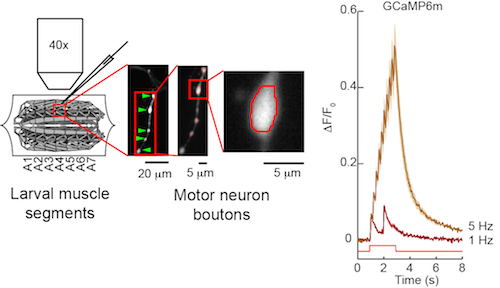
In the Drosophila larval neuromuscular junction assay, motor neurons are directly stimulated using a suction electrode, and responses of boutons are monitored using widefield imaging. Motor neurons can be driven to high frequencies and thus are useful for identifying high-speed sensors for fly optical neurophysiology.
The fly larval neuromuscular junction is used to test sensors in insect neurons. Motor neurons expressing a sensor such as GCaMP6m are directly stimulated and sensor responses in motor neurons are imaged (Chen et al., 2013).
Fly visual system assay
We image optic lobe responses in fly lamina and lobula plate neurons with visual stimuli using two-photon microscopy. We are also developing an assay for spiking interneurons in the optic lobe. Visual system neurons allow us to test sensors with precise stimulus control and high dynamic range of neuronal activity.
Mouse visual cortex assay
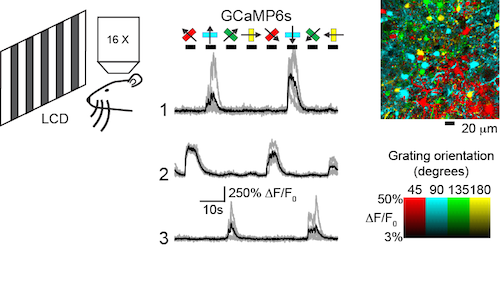
In the mouse visual cortex assay, sensor performance is assayed in hundreds of neurons simultaneously after long-term expression. Neurons in various layers are imaged using two-photon microscopy while drifting gratings are presented to the animal. Virtually all neurons can be driven to spike, and individual neurons exhibit well-established tuning (orientation- and direction-selectivity). Possible cytopathic effects of long-term expression should be detectable as a run-down of this exquisite tuning.
Sensor performance is tested in mouse visual cortical neurons using visual stimuli of drifting gratings at different orientations and two-photon microscopy (left). Orientation-tuned responses of three neurons expressing GCaMP6s are shown (middle). A pixel-based orientation map of GCaMP6s responses (right) reveals the orientation tuning of somata and processes (Chen et al., 2013).
Accomplishments
- Established neuron-based, stimulation and imaging platform for sensor screening
- Engineered GCaMP6 family of ultrasensitive calcium indicators for neuronal population imaging
- Optimized jRCaMP1 and jRGECO1 family of red calcium indicators for deep, dual-color, and channelrhodopsin-2-compatible neuronal activity imaging
- Developed light-gated integrator of calcium signaling, CaMPARI
- Optimized ratiometric, FRET-based calcium sensor, Twitch
Understanding the function and plasticity of neural circuits requires measurements of neural activity over a large range of spatial and temporal scales. We are engineering fluorescent sensors for imaging of neuronal activity in the intact nervous system of experimental animals.
Research proposals
For more details see: