About the Data
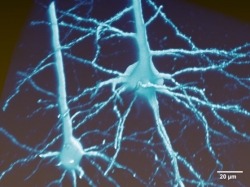
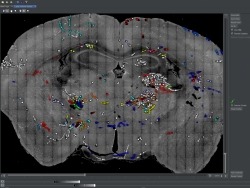
The MouseLight project generates single neuron reconstructions of complete axonal arbors from light microscopic image data in sparsely labeled brain samples. Approximately 100 neurons are reconstructed in every sample, individual brains are registered to the Allen Reference Atlas and the traced neurons are presented in this reference space. The reconstructions included in the MouseLight database were all done by a team of trained annotators. All software and documentation can be found on our github page.
Janelia Workstation
A neuroscience discovery platform that supporting processing, analysis, and annotation of large-scale 3d microscopy data. The MouseLight tools are fully open sourced and made available here with complete documentation. These tools enable semi-automated neuron tracing on terabyte-scale Mouse brain volumes in both 2d and 3d.
MouseLight Acquisition Pipeline
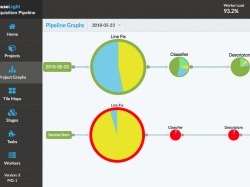
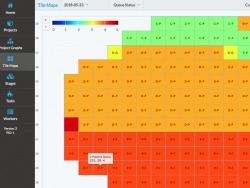
The Mouse Light Pipeline is a collection of services that facilitate the multi-step data processing of the several-thousand tiles per acquisition sample. The processing may occur in real-time as the data becomes available, offline after acquisition is complete, or a combination of both. Each stage of processing can be customized typically through a shell script that calls a function written in Python, MATLAB, or any other language or tool. Different stages can be daisy chained together to form new processing pipelines. Compute resources may be added or removed from the system at any time as well as configured to take on more or less processing load.
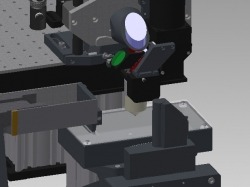
MouseLight Microscope
Hardware plans, component and assembly guide for the MouseLight microscope system are available here. The microscope is controlled using an open source software package called Fetch. This program was developed to image an entire mouse brain by alternating between imaging, and removing the top brain layer using an integrated vibratome.