Filter
Associated Lab
- Dudman Lab (1) Apply Dudman Lab filter
- Hermundstad Lab (1) Apply Hermundstad Lab filter
- Lavis Lab (1) Apply Lavis Lab filter
- Lee (Albert) Lab (2) Apply Lee (Albert) Lab filter
- Looger Lab (2) Apply Looger Lab filter
- Pachitariu Lab (1) Apply Pachitariu Lab filter
- Saalfeld Lab (2) Apply Saalfeld Lab filter
- Scheffer Lab (1) Apply Scheffer Lab filter
- Spruston Lab (1) Apply Spruston Lab filter
- Remove Sternson Lab filter Sternson Lab
- Svoboda Lab (4) Apply Svoboda Lab filter
- Tillberg Lab (3) Apply Tillberg Lab filter
- Turaga Lab (1) Apply Turaga Lab filter
Associated Project Team
Associated Support Team
- Integrative Imaging (2) Apply Integrative Imaging filter
- Janelia Experimental Technology (4) Apply Janelia Experimental Technology filter
- Molecular Genomics (1) Apply Molecular Genomics filter
- Scientific Computing Software (1) Apply Scientific Computing Software filter
- Vivarium (1) Apply Vivarium filter
Publication Date
- 2023 (2) Apply 2023 filter
- 2022 (3) Apply 2022 filter
- 2021 (3) Apply 2021 filter
- 2020 (4) Apply 2020 filter
- 2019 (2) Apply 2019 filter
- 2017 (2) Apply 2017 filter
- 2016 (3) Apply 2016 filter
- 2015 (7) Apply 2015 filter
- 2014 (5) Apply 2014 filter
- 2013 (3) Apply 2013 filter
- 2012 (4) Apply 2012 filter
- 2011 (6) Apply 2011 filter
- 2010 (1) Apply 2010 filter
- 2009 (1) Apply 2009 filter
- 2008 (1) Apply 2008 filter
52 Results
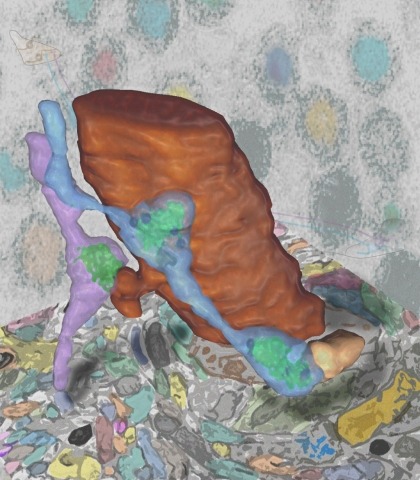
Showing 1-10 of 52 resultsSynaptic connectivity and molecular composition provide a blueprint for information processing in neural circuits. Detailed structural analysis of neural circuits requires nanometer resolution, which can be obtained with serial-section electron microscopy. However, this technique remains challenging for reconstructing molecularly defined synapses. We used a genetically encoded synaptic marker for electron microscopy (GESEM) based on intra-vesicular generation of electron-dense labeling in axonal boutons. This approach allowed the identification of synapses from Cre recombinase-expressing or GAL4-expressing neurons in the mouse and fly with excellent preservation of ultrastructure. We applied this tool to visualize long-range connectivity of AGRP and POMC neurons in the mouse, two molecularly defined hypothalamic populations that are important for feeding behavior. Combining selective ultrastructural reconstruction of neuropil with functional and viral circuit mapping, we characterized some basic features of circuit organization for axon projections of these cell types. Our findings demonstrate that GESEM labeling enables long-range connectomics with molecularly defined cell types.
Understanding the structure and function of neural circuits are central questions in neuroscience research. To address these questions, new genetically encoded tools have been developed for mapping, monitoring, and manipulating neurons. Essential to implementation of these tools is their selective delivery to defined neuronal populations in the brain. This has been facilitated by recent improvements in cell type-specific transgene expression using recombinant adeno-associated viral vectors. Here, we highlight these developments and discuss areas for improvement that could further expand capabilities for neural circuit analysis.
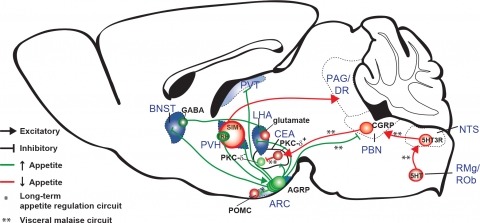
New tools for mapping and manipulating molecularly defined neural circuits have improved understanding of how the central nervous system regulates appetite. Studies focused on AGRP neurons, a starvation-sensitive hypothalamic population, have identified multiple circuit elements that can elicit or suppress feeding behavior. Distinct axon projections of this neuron population point to different circuits that regulate long-term appetite, short-term feeding, or visceral malaise-mediated anorexia. Here, we review recent studies examining these neural circuits that control food intake. © 2014 S. Karger AG, Basel.
Two intermingled hypothalamic neuron populations specified by expression of agouti-related peptide (AGRP) or pro-opiomelanocortin (POMC) positively and negatively influence feeding behavior, respectively, possibly by reciprocally regulating downstream melanocortin receptors. However, the sufficiency of these neurons to control behavior and the relationship of their activity to the magnitude and dynamics of feeding are unknown. To measure this, we used channelrhodopsin-2 for cell type-specific photostimulation. Activation of only 800 AGRP neurons in mice evoked voracious feeding within minutes. The behavioral response increased with photoexcitable neuron number, photostimulation frequency and stimulus duration. Conversely, POMC neuron stimulation reduced food intake and body weight, which required melanocortin receptor signaling. However, AGRP neuron-mediated feeding was not dependent on suppressing this melanocortin pathway, indicating that AGRP neurons directly engage feeding circuits. Furthermore, feeding was evoked selectively over drinking without training or prior photostimulus exposure, which suggests that AGRP neurons serve a dedicated role coordinating this complex behavior.
Advances in neuro-technology for mapping, manipulating, and monitoring molecularly defined cell types are rapidly advancing insight into neural circuits that regulate appetite. Here, we review these important tools and their applications in circuits that control food seeking and consumption. Technical capabilities provided by these tools establish a rigorous experimental framework for research into the neurobiology of hunger.
The nucleus accumbens regulates consummatory behaviors, such as eating. In this issue of Neuron, O'Connor et al. (2015) identify dopamine receptor 1-expressing neurons that project to the lateral hypothalamus as mediating rapid control over feeding behavior.
Digital reconstruction of 3D neuron structures is an important step toward reverse engineering the wiring and functions of a brain. However, despite a number of existing studies, this task is still challenging, especially when a 3D microscopic image has low single-to-noise ratio and discontinued segments of neurite patterns.
Brains encode behaviors using neurons amenable to systematic classification by gene expression. The contribution of molecular identity to neural coding is not understood because of the challenges involved with measuring neural dynamics and molecular information from the same cells. We developed CaRMA (calcium and RNA multiplexed activity) imaging based on recording in vivo single-neuron calcium dynamics followed by gene expression analysis. We simultaneously monitored activity in hundreds of neurons in mouse paraventricular hypothalamus (PVH). Combinations of cell-type marker genes had predictive power for neuronal responses across 11 behavioral states. The PVH uses combinatorial assemblies of molecularly defined neuron populations for grouped-ensemble coding of survival behaviors. The neuropeptide receptor neuropeptide Y receptor type 1 (Npy1r) amalgamated multiple cell types with similar responses. Our results show that molecularly defined neurons are important processing units for brain function.
N-Methyl-D-aspartate receptors (NMDA-Rs) are ion channels that are important for synaptic plasticity, which is involved in learning and drug addiction. We show enzymatic targeting of an NMDA-R antagonist, MK801, to a molecularly defined neuronal population with the cell-type-selectivity of genetic methods and the temporal control of pharmacology. We find that NMDA-Rs on dopamine neurons are necessary for cocaine-induced synaptic potentiation, demonstrating that cell type-specific pharmacology can be used to dissect signaling pathways within complex brain circuits.